Aimin Pan
FlattenQuant: Breaking Through the Inference Compute-bound for Large Language Models with Per-tensor Quantization
Feb 28, 2024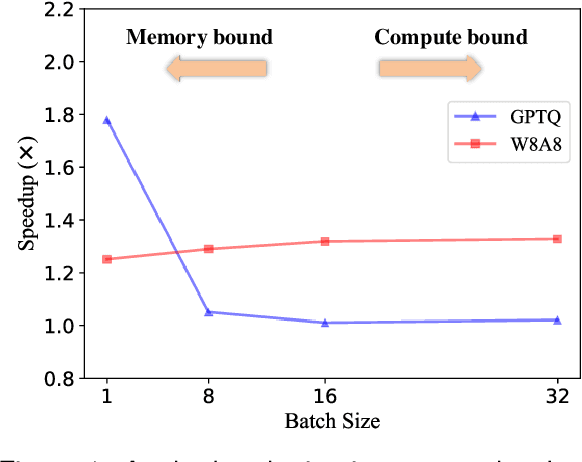
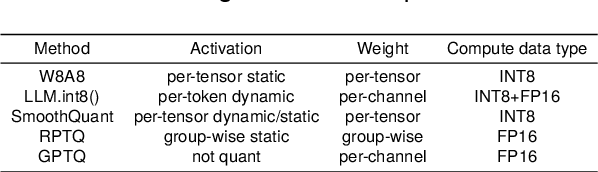
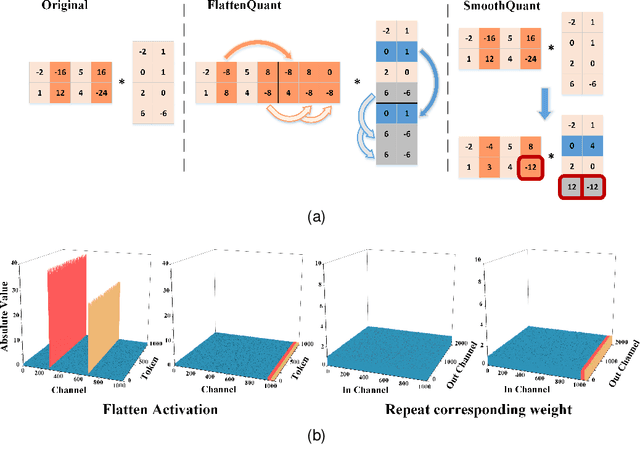
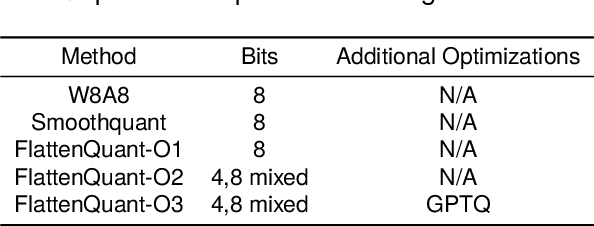
Abstract:Large language models (LLMs) have demonstrated state-of-the-art performance across various tasks. However, the latency of inference and the large GPU memory consumption of LLMs restrict their deployment performance. Recently, there have been some efficient attempts to quantize LLMs, yet inference with large batch size or long sequence still has the issue of being compute-bound. Fine-grained quantization methods have showcased their proficiency in achieving low-bit quantization for LLMs, while requiring FP16 data type for linear layer computations, which is time-consuming when dealing with large batch size or long sequence. In this paper, we introduce a method called FlattenQuant, which significantly reduces the maximum value of the tensor by flattening the large channels in the tensor, to achieve low bit per-tensor quantization with minimal accuracy loss. Our experiments show that FlattenQuant can directly use 4 bits to achieve 48.29% of the linear layer calculation in LLMs, with the remaining layers using 8 bits. The 4-bit matrix multiplication introduced in the FlattenQuant method can effectively address the compute-bound caused by large matrix calculation. Our work achieves up to 2$\times$ speedup and 2.3$\times$ memory reduction for LLMs with negligible loss in accuracy.
Holmes: Towards Distributed Training Across Clusters with Heterogeneous NIC Environment
Dec 11, 2023



Abstract:Large language models (LLMs) such as GPT-3, OPT, and LLaMA have demonstrated remarkable accuracy in a wide range of tasks. However, training these models can incur significant expenses, often requiring tens of thousands of GPUs for months of continuous operation. Typically, this training is carried out in specialized GPU clusters equipped with homogeneous high-speed Remote Direct Memory Access (RDMA) network interface cards (NICs). The acquisition and maintenance of such dedicated clusters is challenging. Current LLM training frameworks, like Megatron-LM and Megatron-DeepSpeed, focus primarily on optimizing training within homogeneous cluster settings. In this paper, we introduce Holmes, a training framework for LLMs that employs thoughtfully crafted data and model parallelism strategies over the heterogeneous NIC environment. Our primary technical contribution lies in a novel scheduling method that intelligently allocates distinct computational tasklets in LLM training to specific groups of GPU devices based on the characteristics of their connected NICs. Furthermore, our proposed framework, utilizing pipeline parallel techniques, demonstrates scalability to multiple GPU clusters, even in scenarios without high-speed interconnects between nodes in distinct clusters. We conducted comprehensive experiments that involved various scenarios in the heterogeneous NIC environment. In most cases, our framework achieves performance levels close to those achievable with homogeneous RDMA-capable networks (InfiniBand or RoCE), significantly exceeding training efficiency within the pure Ethernet environment. Additionally, we verified that our framework outperforms other mainstream LLM frameworks under heterogeneous NIC environment in terms of training efficiency and can be seamlessly integrated with them.
Exploring Post-Training Quantization of Protein Language Models
Oct 30, 2023



Abstract:Recent advancements in unsupervised protein language models (ProteinLMs), like ESM-1b and ESM-2, have shown promise in different protein prediction tasks. However, these models face challenges due to their high computational demands, significant memory needs, and latency, restricting their usage on devices with limited resources. To tackle this, we explore post-training quantization (PTQ) for ProteinLMs, focusing on ESMFold, a simplified version of AlphaFold based on ESM-2 ProteinLM. Our study is the first attempt to quantize all weights and activations of ProteinLMs. We observed that the typical uniform quantization method performs poorly on ESMFold, causing a significant drop in TM-Score when using 8-bit quantization. We conducted extensive quantization experiments, uncovering unique challenges associated with ESMFold, particularly highly asymmetric activation ranges before Layer Normalization, making representation difficult using low-bit fixed-point formats. To address these challenges, we propose a new PTQ method for ProteinLMs, utilizing piecewise linear quantization for asymmetric activation values to ensure accurate approximation. We demonstrated the effectiveness of our method in protein structure prediction tasks, demonstrating that ESMFold can be accurately quantized to low-bit widths without compromising accuracy. Additionally, we applied our method to the contact prediction task, showcasing its versatility. In summary, our study introduces an innovative PTQ method for ProteinLMs, addressing specific quantization challenges and potentially leading to the development of more efficient ProteinLMs with significant implications for various protein-related applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge