Adam Gonczarek
Similarity-based Memory Enhanced Joint Entity and Relation Extraction
Jul 14, 2023Abstract:Document-level joint entity and relation extraction is a challenging information extraction problem that requires a unified approach where a single neural network performs four sub-tasks: mention detection, coreference resolution, entity classification, and relation extraction. Existing methods often utilize a sequential multi-task learning approach, in which the arbitral decomposition causes the current task to depend only on the previous one, missing the possible existence of the more complex relationships between them. In this paper, we present a multi-task learning framework with bidirectional memory-like dependency between tasks to address those drawbacks and perform the joint problem more accurately. Our empirical studies show that the proposed approach outperforms the existing methods and achieves state-of-the-art results on the BioCreative V CDR corpus.
Classical Out-of-Distribution Detection Methods Benchmark in Text Classification Tasks
Jul 13, 2023
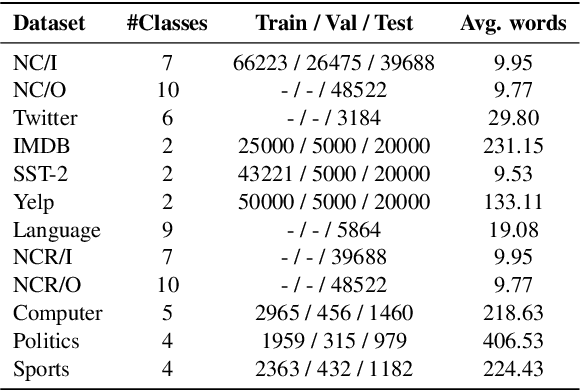
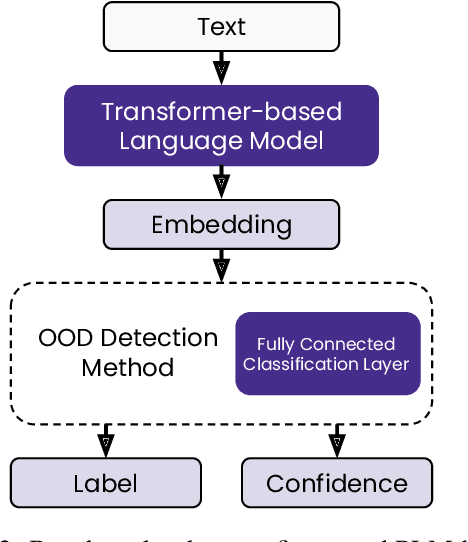
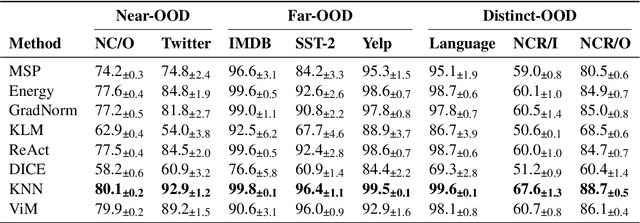
Abstract:State-of-the-art models can perform well in controlled environments, but they often struggle when presented with out-of-distribution (OOD) examples, making OOD detection a critical component of NLP systems. In this paper, we focus on highlighting the limitations of existing approaches to OOD detection in NLP. Specifically, we evaluated eight OOD detection methods that are easily integrable into existing NLP systems and require no additional OOD data or model modifications. One of our contributions is providing a well-structured research environment that allows for full reproducibility of the results. Additionally, our analysis shows that existing OOD detection methods for NLP tasks are not yet sufficiently sensitive to capture all samples characterized by various types of distributional shifts. Particularly challenging testing scenarios arise in cases of background shift and randomly shuffled word order within in domain texts. This highlights the need for future work to develop more effective OOD detection approaches for the NLP problems, and our work provides a well-defined foundation for further research in this area.
Domain-Agnostic Neural Architecture for Class Incremental Continual Learning in Document Processing Platform
Jul 11, 2023Abstract:Production deployments in complex systems require ML architectures to be highly efficient and usable against multiple tasks. Particularly demanding are classification problems in which data arrives in a streaming fashion and each class is presented separately. Recent methods with stochastic gradient learning have been shown to struggle in such setups or have limitations like memory buffers, and being restricted to specific domains that disable its usage in real-world scenarios. For this reason, we present a fully differentiable architecture based on the Mixture of Experts model, that enables the training of high-performance classifiers when examples from each class are presented separately. We conducted exhaustive experiments that proved its applicability in various domains and ability to learn online in production environments. The proposed technique achieves SOTA results without a memory buffer and clearly outperforms the reference methods.
Neural Architecture for Online Ensemble Continual Learning
Nov 27, 2022Abstract:Continual learning with an increasing number of classes is a challenging task. The difficulty rises when each example is presented exactly once, which requires the model to learn online. Recent methods with classic parameter optimization procedures have been shown to struggle in such setups or have limitations like non-differentiable components or memory buffers. For this reason, we present the fully differentiable ensemble method that allows us to efficiently train an ensemble of neural networks in the end-to-end regime. The proposed technique achieves SOTA results without a memory buffer and clearly outperforms the reference methods. The conducted experiments have also shown a significant increase in the performance for small ensembles, which demonstrates the capability of obtaining relatively high classification accuracy with a reduced number of classifiers.
Towards fully automated protein structure elucidation with NMR spectroscopy
Jul 31, 2018
Abstract:Nuclear magnetic resonance (NMR) spectroscopy is one of the leading techniques for protein studies. The method features a number of properties, allowing to explain macromolecular interactions mechanistically and resolve structures with atomic resolution. However, due to laborious data analysis, a full potential of NMR spectroscopy remains unexploited. Here we present an approach aiming at automation of two major bottlenecks in the analysis pipeline, namely, peak picking and chemical shift assignment. Our approach combines deep learning, non-parametric models and combinatorial optimization, and is able to detect signals of interest in a multidimensional NMR data with high accuracy and match them with atoms in medium-length protein sequences, which is a preliminary step to solve protein spatial structure.
Learning Deep Architectures for Interaction Prediction in Structure-based Virtual Screening
Sep 19, 2017
Abstract:We introduce a deep learning architecture for structure-based virtual screening that generates fixed-sized fingerprints of proteins and small molecules by applying learnable atom convolution and softmax operations to each compound separately. These fingerprints are further transformed non-linearly, their inner-product is calculated and used to predict the binding potential. Moreover, we show that widely used benchmark datasets may be insufficient for testing structure-based virtual screening methods that utilize machine learning. Therefore, we introduce a new benchmark dataset, which we constructed based on DUD-E and PDBBind databases.
Subspace Restricted Boltzmann Machine
Jul 16, 2014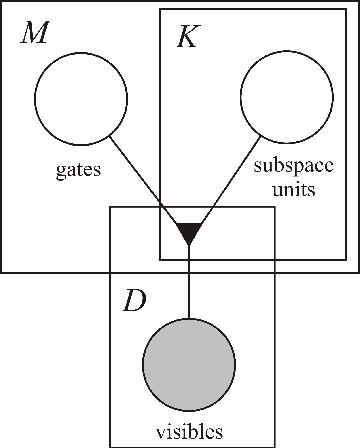
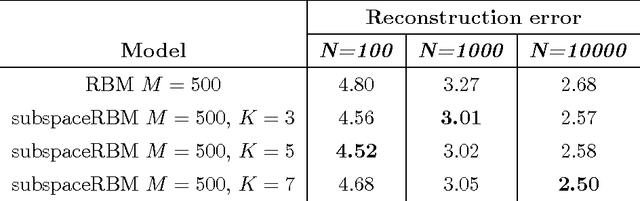

Abstract:The subspace Restricted Boltzmann Machine (subspaceRBM) is a third-order Boltzmann machine where multiplicative interactions are between one visible and two hidden units. There are two kinds of hidden units, namely, gate units and subspace units. The subspace units reflect variations of a pattern in data and the gate unit is responsible for activating the subspace units. Additionally, the gate unit can be seen as a pooling feature. We evaluate the behavior of subspaceRBM through experiments with MNIST digit recognition task, measuring reconstruction error and classification error.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge