Abdulkadir Gokce
Contour Integration Underlies Human-Like Vision
Apr 07, 2025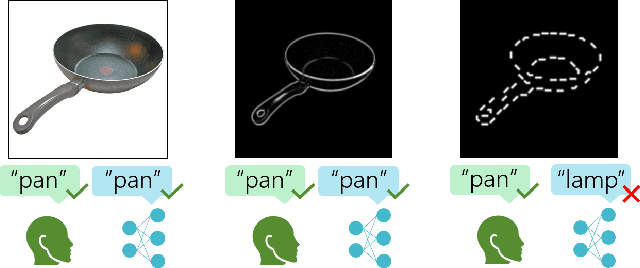
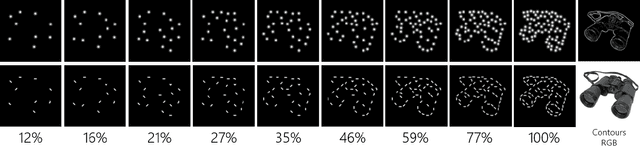
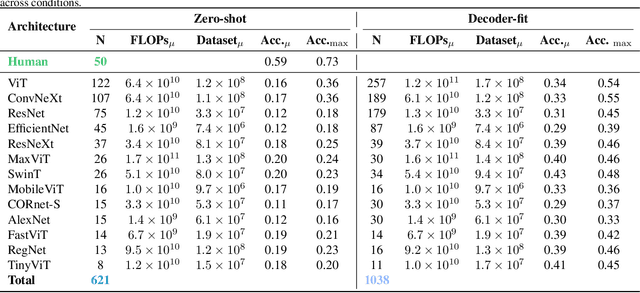
Abstract:Despite the tremendous success of deep learning in computer vision, models still fall behind humans in generalizing to new input distributions. Existing benchmarks do not investigate the specific failure points of models by analyzing performance under many controlled conditions. Our study systematically dissects where and why models struggle with contour integration -- a hallmark of human vision -- by designing an experiment that tests object recognition under various levels of object fragmentation. Humans (n=50) perform at high accuracy, even with few object contours present. This is in contrast to models which exhibit substantially lower sensitivity to increasing object contours, with most of the over 1,000 models we tested barely performing above chance. Only at very large scales ($\sim5B$ training dataset size) do models begin to approach human performance. Importantly, humans exhibit an integration bias -- a preference towards recognizing objects made up of directional fragments over directionless fragments. We find that not only do models that share this property perform better at our task, but that this bias also increases with model training dataset size, and training models to exhibit contour integration leads to high shape bias. Taken together, our results suggest that contour integration is a hallmark of object vision that underlies object recognition performance, and may be a mechanism learned from data at scale.
Scaling Laws for Task-Optimized Models of the Primate Visual Ventral Stream
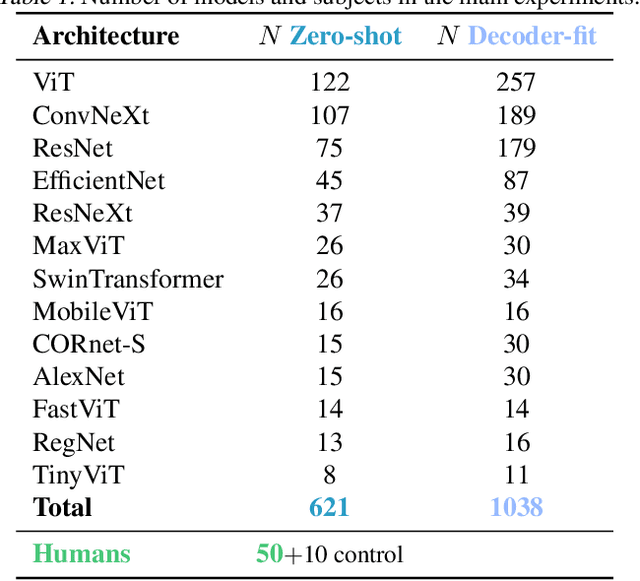
Nov 08, 2024Abstract:When trained on large-scale object classification datasets, certain artificial neural network models begin to approximate core object recognition (COR) behaviors and neural response patterns in the primate visual ventral stream (VVS). While recent machine learning advances suggest that scaling model size, dataset size, and compute resources improve task performance, the impact of scaling on brain alignment remains unclear. In this study, we explore scaling laws for modeling the primate VVS by systematically evaluating over 600 models trained under controlled conditions on benchmarks spanning V1, V2, V4, IT and COR behaviors. We observe that while behavioral alignment continues to scale with larger models, neural alignment saturates. This observation remains true across model architectures and training datasets, even though models with stronger inductive bias and datasets with higher-quality images are more compute-efficient. Increased scaling is especially beneficial for higher-level visual areas, where small models trained on few samples exhibit only poor alignment. Finally, we develop a scaling recipe, indicating that a greater proportion of compute should be allocated to data samples over model size. Our results suggest that while scaling alone might suffice for alignment with human core object recognition behavior, it will not yield improved models of the brain's visual ventral stream with current architectures and datasets, highlighting the need for novel strategies in building brain-like models.
EndoL2H: Deep Super-Resolution for Capsule Endoscopy
Feb 13, 2020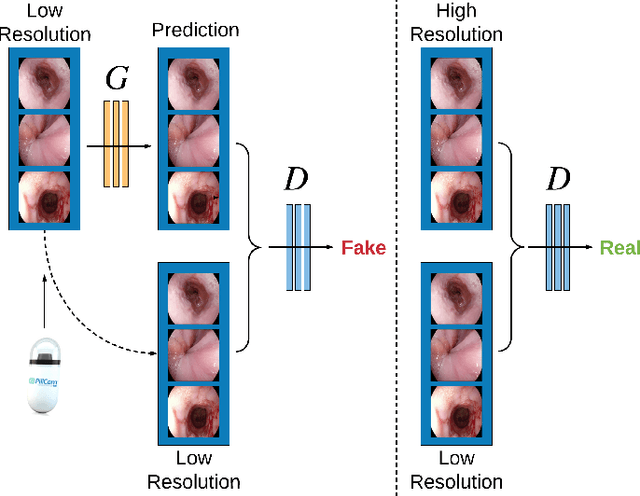
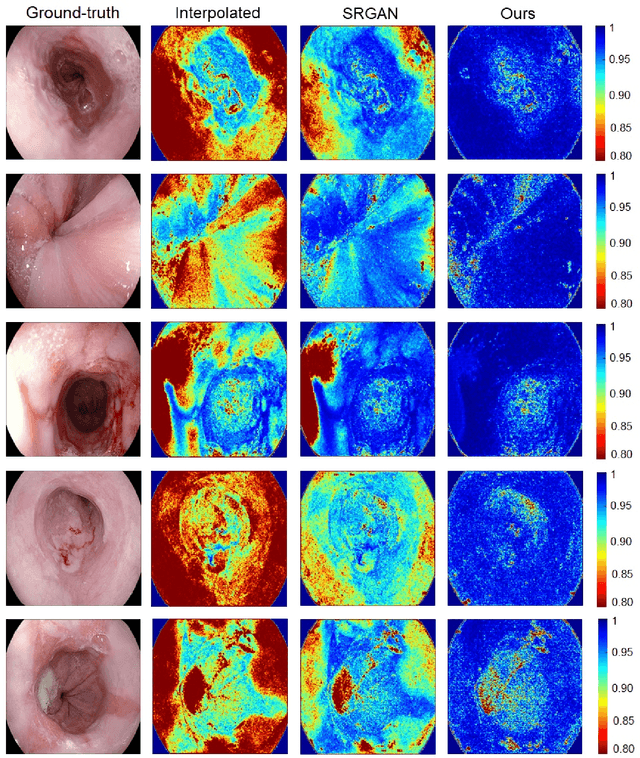
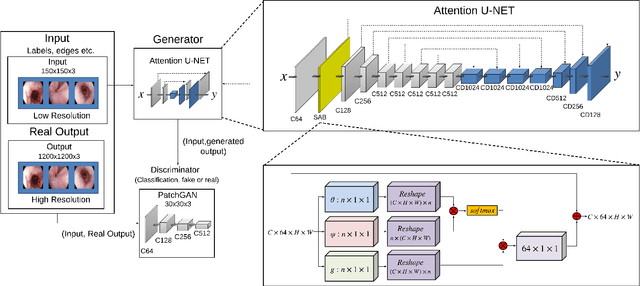

Abstract:Wireless capsule endoscopy is the preferred modality for diagnosis and assessment of small bowel disease. However, the poor resolution is a limitation for both subjective and automated diagnostics. Enhanced-resolution endoscopy has shown to improve adenoma detection rate for conventional endoscopy and is likely to do the same for capsule endoscopy. In this work, we propose and quantitatively validate a novel framework to learn a mapping from low-to-high resolution endoscopic images. We use conditional adversarial networks and spatial attention to improve the resolution by up to a factor of 8x. Our quantitative study demonstrates the superiority of our proposed approach over Super-Resolution Generative Adversarial Network (SRGAN) and bicubic interpolation. For qualitative analysis, visual Turing tests were performed by 16 gastroenterologists to confirm the clinical utility of the proposed approach. Our approach is generally applicable to any endoscopic capsule system and has the potential to improve diagnosis and better harness computational approaches for polyp detection and characterization. Our code and trained models are available at https://github.com/akgokce/EndoL2H.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge