Topology-Preserving Shape Reconstruction and Registration via Neural Diffeomorphic Flow
Paper and Code
Mar 21, 2022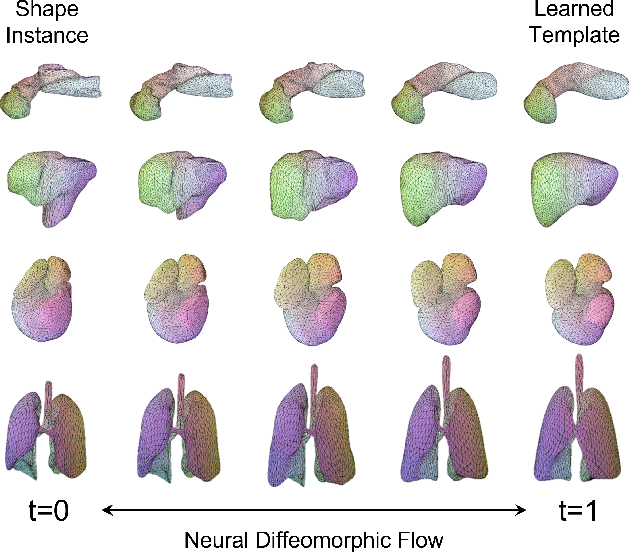
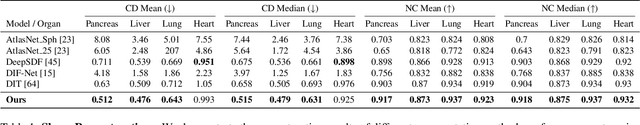
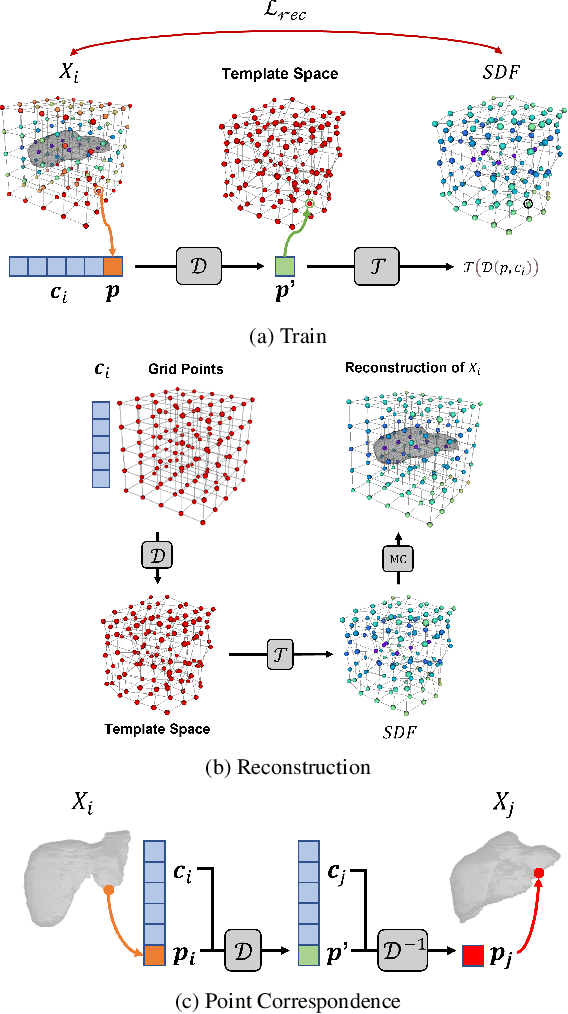
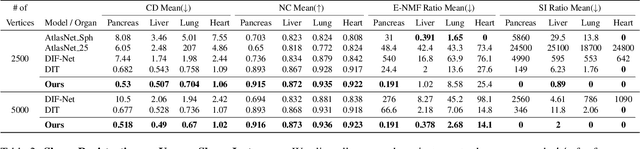
Deep Implicit Functions (DIFs) represent 3D geometry with continuous signed distance functions learned through deep neural nets. Recently DIFs-based methods have been proposed to handle shape reconstruction and dense point correspondences simultaneously, capturing semantic relationships across shapes of the same class by learning a DIFs-modeled shape template. These methods provide great flexibility and accuracy in reconstructing 3D shapes and inferring correspondences. However, the point correspondences built from these methods do not intrinsically preserve the topology of the shapes, unlike mesh-based template matching methods. This limits their applications on 3D geometries where underlying topological structures exist and matter, such as anatomical structures in medical images. In this paper, we propose a new model called Neural Diffeomorphic Flow (NDF) to learn deep implicit shape templates, representing shapes as conditional diffeomorphic deformations of templates, intrinsically preserving shape topologies. The diffeomorphic deformation is realized by an auto-decoder consisting of Neural Ordinary Differential Equation (NODE) blocks that progressively map shapes to implicit templates. We conduct extensive experiments on several medical image organ segmentation datasets to evaluate the effectiveness of NDF on reconstructing and aligning shapes. NDF achieves consistently state-of-the-art organ shape reconstruction and registration results in both accuracy and quality. The source code is publicly available at https://github.com/Siwensun/Neural_Diffeomorphic_Flow--NDF.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge