Structured Q-learning For Antibody Design
Paper and Code
Sep 13, 2022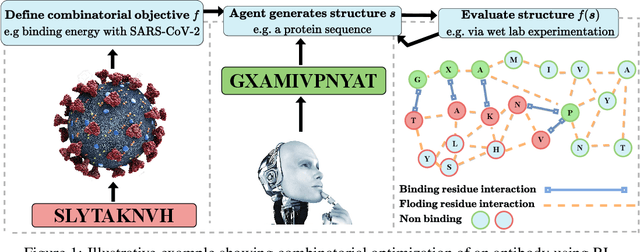
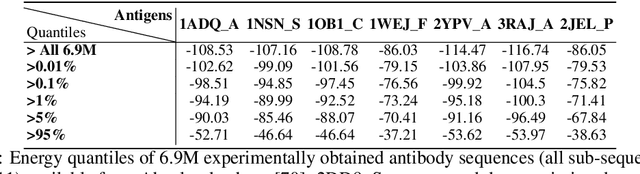
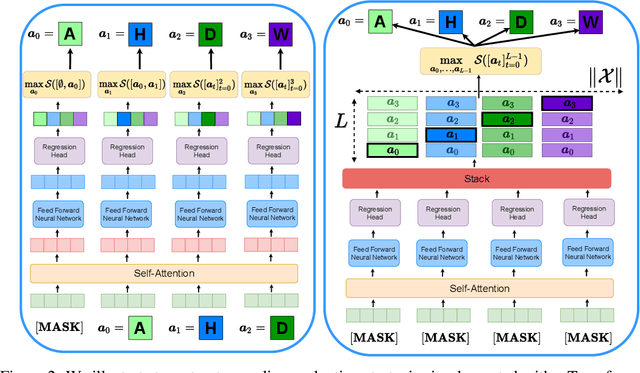
Optimizing combinatorial structures is core to many real-world problems, such as those encountered in life sciences. For example, one of the crucial steps involved in antibody design is to find an arrangement of amino acids in a protein sequence that improves its binding with a pathogen. Combinatorial optimization of antibodies is difficult due to extremely large search spaces and non-linear objectives. Even for modest antibody design problems, where proteins have a sequence length of eleven, we are faced with searching over 2.05 x 10^14 structures. Applying traditional Reinforcement Learning algorithms such as Q-learning to combinatorial optimization results in poor performance. We propose Structured Q-learning (SQL), an extension of Q-learning that incorporates structural priors for combinatorial optimization. Using a molecular docking simulator, we demonstrate that SQL finds high binding energy sequences and performs favourably against baselines on eight challenging antibody design tasks, including designing antibodies for SARS-COV.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge