Multi-Label Robust Factorization Autoencoder and its Application in Predicting Drug-Drug Interactions
Paper and Code
Nov 01, 2018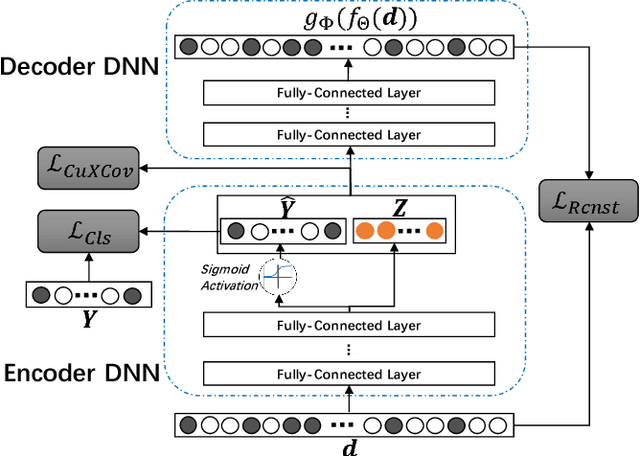
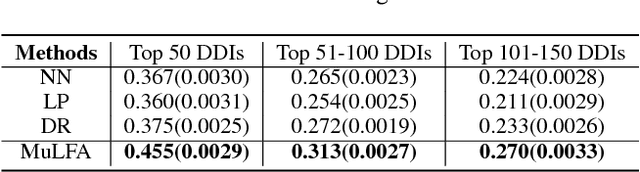
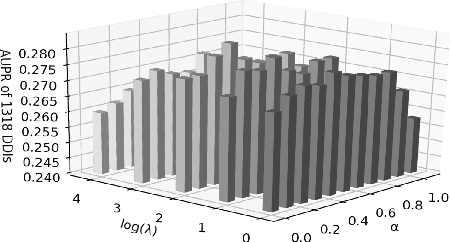

Drug-drug interactions (DDIs) are a major cause of preventable hospitalizations and deaths. Predicting the occurrence of DDIs helps drug safety professionals allocate investigative resources and take appropriate regulatory action promptly. Traditional DDI prediction methods predict DDIs based on the similarity between drugs. Recently, researchers revealed that predictive performance can be improved by better modeling the interactions between drug pairs with bilinear forms. However, the shallow models leveraging bilinear forms suffer from limitations on capturing complicated nonlinear interactions between drug pairs. To this end, we propose Multi-Label Robust Factorization Autoencoder (abbreviated to MuLFA) for DDI prediction, which learns a representation of interactions between drug pairs and has the capability of characterizing complicated nonlinear interactions more precisely. Moreover, a novel loss called CuXCov is designed to effectively learn the parameters of MuLFA. Furthermore, the decoder is able to generate high-risk chemical structures of drug pairs for specific DDIs, assisting pharmacists to better understand the relationship between drug chemistry and DDI. Experimental results on real-world datasets demonstrate that MuLFA consistently outperforms state-of-the-art methods; particularly, it increases 21:3% predictive performance compared to the best baseline for top 50 frequent DDIs.We also illustrate various case studies to demonstrate the efficacy of the chemical structures generated by MuLFA in DDI diagnosis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge