Hierarchical Large-scale Graph Similarity Computation via Graph Coarsening and Matching
Paper and Code
Jun 09, 2020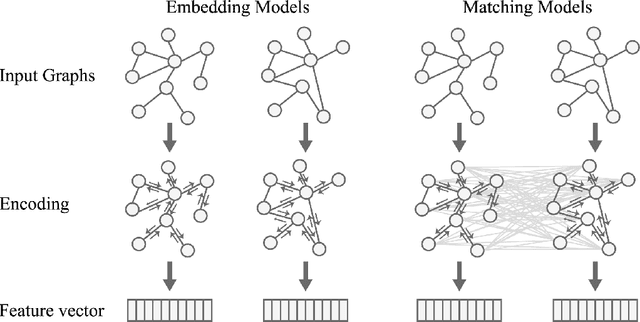
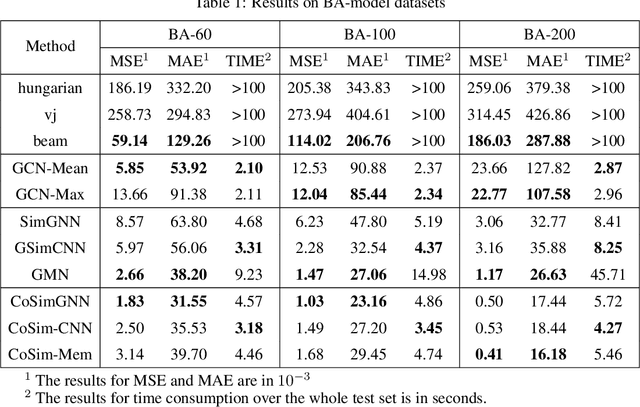
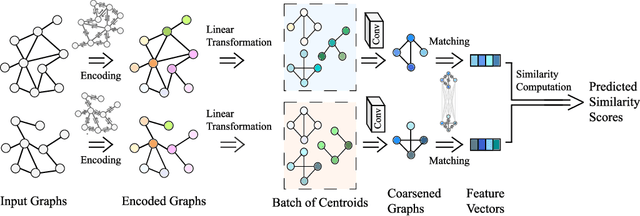
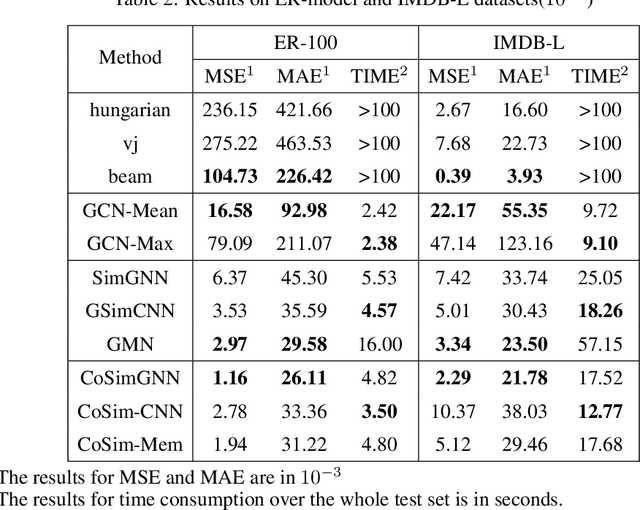
In this work, we focus on large graph similarity computation problem and propose a novel ``embedding-coarsening-matching'' learning framework, which outperforms state-of-the-art methods in this task and has significant improvement in time efficiency. Graph similarity computation for metrics such as Graph Edit Distance (GED) is typically NP-hard, and existing heuristics-based algorithms usually achieves a unsatisfactory trade-off between accuracy and efficiency. Recently the development of deep learning techniques provides a promising solution for this problem by a data-driven approach which trains a network to encode graphs to their own feature vectors and computes similarity based on feature vectors. These deep-learning methods can be classified to two categories, embedding models and matching models. Embedding models such as GCN-Mean and GCN-Max, which directly map graphs to respective feature vectors, run faster but the performance is usually poor due to the lack of interactions across graphs. Matching models such as GMN, whose encoding process involves interaction across the two graphs, are more accurate but interaction between whole graphs brings a significant increase in time consumption (at least quadratic time complexity over number of nodes). Inspired by large biological molecular identification where the whole molecular is first mapped to functional groups and then identified based on these functional groups, our ``embedding-coarsening-matching'' learning framework first embeds and coarsens large graphs to coarsened graphs with denser local topology and then matching mechanism is deployed on the coarsened graphs for the final similarity scores. Detailed experiments have been conducted and the results demonstrate the efficiency and effectiveness of our proposed framework.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge