Domain-Specific Language Model Pretraining for Biomedical Natural Language Processing
Paper and Code
Aug 20, 2020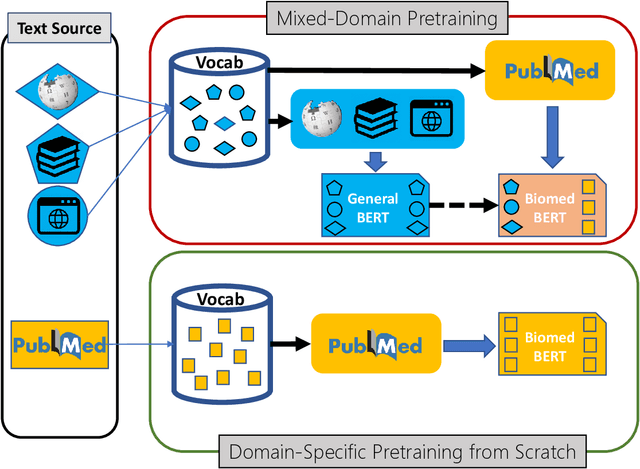
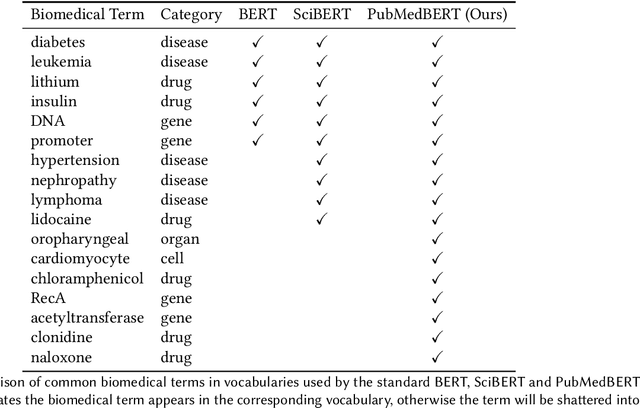
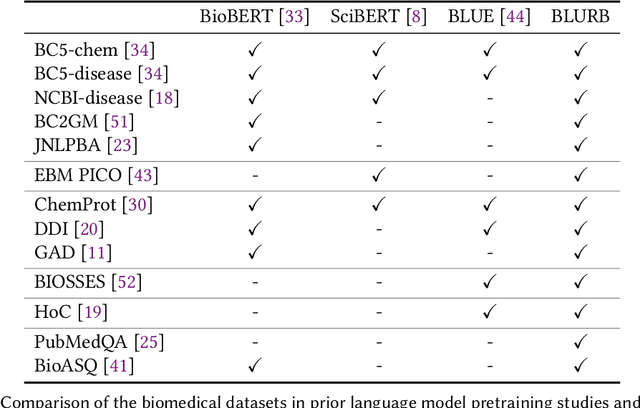
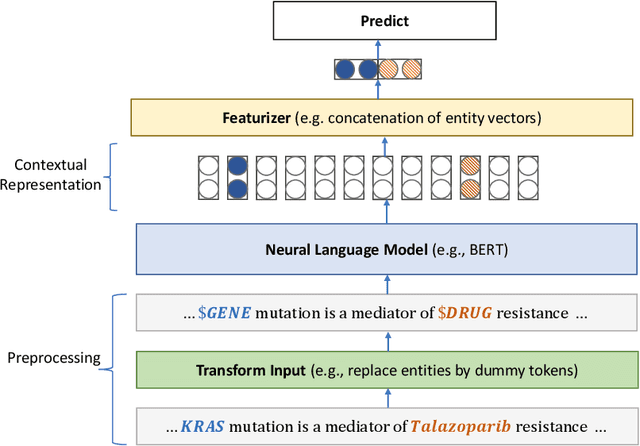
Pretraining large neural language models, such as BERT, has led to impressive gains on many natural language processing (NLP) tasks. However, most pretraining efforts focus on general domain corpora, such as newswire and Web. A prevailing assumption is that even domain-specific pretraining can benefit by starting from general-domain language models. In this paper, we challenge this assumption by showing that for domains with abundant unlabeled text, such as biomedicine, pretraining language models from scratch results in substantial gains over continual pretraining of general-domain language models. To facilitate this investigation, we compile a comprehensive biomedical NLP benchmark from publicly-available datasets. Our experiments show that domain-specific pretraining serves as a solid foundation for a wide range of biomedical NLP tasks, leading to new state-of-the-art results across the board. Further, in conducting a thorough evaluation of modeling choices, both for pretraining and task-specific fine-tuning, we discover that some common practices are unnecessary with BERT models, such as using complex tagging schemes in named entity recognition (NER). To help accelerate research in biomedical NLP, we have released our state-of-the-art pretrained and task-specific models for the community, and created a leaderboard featuring our BLURB benchmark (short for Biomedical Language Understanding & Reasoning Benchmark) at https://aka.ms/BLURB.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge