Zuhui Wang
Enhancing Image-Text Matching with Adaptive Feature Aggregation
Jan 18, 2024Abstract:Image-text matching aims to find matched cross-modal pairs accurately. While current methods often rely on projecting cross-modal features into a common embedding space, they frequently suffer from imbalanced feature representations across different modalities, leading to unreliable retrieval results. To address these limitations, we introduce a novel Feature Enhancement Module that adaptively aggregates single-modal features for more balanced and robust image-text retrieval. Additionally, we propose a new loss function that overcomes the shortcomings of original triplet ranking loss, thereby significantly improving retrieval performance. The proposed model has been evaluated on two public datasets and achieves competitive retrieval performance when compared with several state-of-the-art models. Implementation codes can be found here.
Cross-domain Microscopy Cell Counting by Disentangled Transfer Learning
Nov 26, 2022Abstract:Microscopy cell images of biological experiments on different tissues/organs/imaging conditions usually contain cells with various shapes and appearances on different image backgrounds, making a cell counting model trained in a source domain hard to be transferred to a new target domain. Thus, costly manual annotation is required to train deep learning-based cell counting models across different domains. Instead, we propose a cross-domain cell counting approach with only a little human annotation effort. First, we design a cell counting network that can disentangle domain-specific knowledge and domain-agnostic knowledge in cell images, which are related to the generation of domain style images and cell density maps, respectively. Secondly, we propose an image synthesis method capable of synthesizing a large number of images based on a few annotated ones. Finally, we use a public dataset of synthetic cells, which has no annotation cost at all as the source domain to train our cell counting network; then, only the domain-agnostic knowledge in the trained model is transferred to a new target domain of real cell images, by progressively fine-tuning the trained model using synthesized target-domain images and a few annotated ones. Evaluated on two public target datasets of real cell images, our cross-domain cell counting approach that only needs annotation on a few images in a new target domain achieves good performance, compared to state-of-the-art methods that rely on fully annotated training images in the target domain.
TimeVAE: A Variational Auto-Encoder for Multivariate Time Series Generation
Dec 07, 2021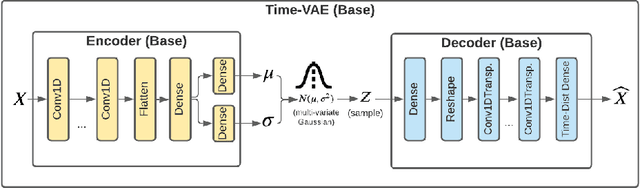



Abstract:Recent work in synthetic data generation in the time-series domain has focused on the use of Generative Adversarial Networks. We propose a novel architecture for synthetically generating time-series data with the use of Variational Auto-Encoders (VAEs). The proposed architecture has several distinct properties: interpretability, ability to encode domain knowledge, and reduced training times. We evaluate data generation quality by similarity and predictability against four multivariate datasets. We experiment with varying sizes of training data to measure the impact of data availability on generation quality for our VAE method as well as several state-of-the-art data generation methods. Our results on similarity tests show that the VAE approach is able to accurately represent the temporal attributes of the original data. On next-step prediction tasks using generated data, the proposed VAE architecture consistently meets or exceeds performance of state-of-the-art data generation methods. While noise reduction may cause the generated data to deviate from original data, we demonstrate the resulting de-noised data can significantly improve performance for next-step prediction using generated data. Finally, the proposed architecture can incorporate domain-specific time-patterns such as polynomial trends and seasonalities to provide interpretable outputs. Such interpretability can be highly advantageous in applications requiring transparency of model outputs or where users desire to inject prior knowledge of time-series patterns into the generative model.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge