Zahra Montazeri
VideoNeuMat: Neural Material Extraction from Generative Video Models
Feb 06, 2026Abstract:Creating photorealistic materials for 3D rendering requires exceptional artistic skill. Generative models for materials could help, but are currently limited by the lack of high-quality training data. While recent video generative models effortlessly produce realistic material appearances, this knowledge remains entangled with geometry and lighting. We present VideoNeuMat, a two-stage pipeline that extracts reusable neural material assets from video diffusion models. First, we finetune a large video model (Wan 2.1 14B) to generate material sample videos under controlled camera and lighting trajectories, effectively creating a "virtual gonioreflectometer" that preserves the model's material realism while learning a structured measurement pattern. Second, we reconstruct compact neural materials from these videos through a Large Reconstruction Model (LRM) finetuned from a smaller Wan 1.3B video backbone. From 17 generated video frames, our LRM performs single-pass inference to predict neural material parameters that generalize to novel viewing and lighting conditions. The resulting materials exhibit realism and diversity far exceeding the limited synthetic training data, demonstrating that material knowledge can be successfully transferred from internet-scale video models into standalone, reusable neural 3D assets.
An Improved NeuMIP with Better Accuracy
Jul 19, 2023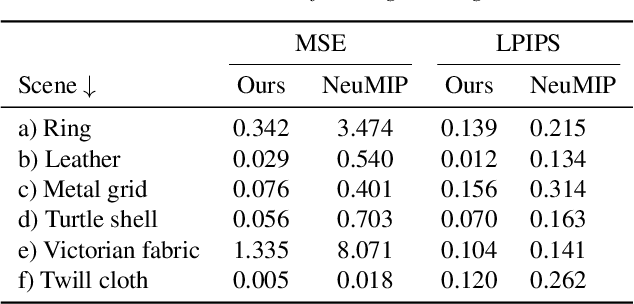



Abstract:Neural reflectance models are capable of accurately reproducing the spatially-varying appearance of many real-world materials at different scales. However, existing methods have difficulties handling highly glossy materials. To address this problem, we introduce a new neural reflectance model which, compared with existing methods, better preserves not only specular highlights but also fine-grained details. To this end, we enhance the neural network performance by encoding input data to frequency space, inspired by NeRF, to better preserve the details. Furthermore, we introduce a gradient-based loss and employ it in multiple stages, adaptive to the progress of the learning phase. Lastly, we utilize an optional extension to the decoder network using the Inception module for more accurate yet costly performance. We demonstrate the effectiveness of our method using a variety of synthetic and real examples.
Learning to Rasterize Differentiable
Nov 23, 2022Abstract:Differentiable rasterization changes the common formulation of primitive rasterization -- which has zero gradients almost everywhere, due to discontinuous edges and occlusion -- to an alternative one, which is not subject to this limitation and has similar optima. These alternative versions in general are ''soft'' versions of the original one. Unfortunately, it is not clear, what exact way of softening will provide the best performance in terms of converging the most reliability to a desired goal. Previous work has analyzed and compared several combinations of softening. In this work, we take it a step further and, instead of making a combinatorical choice of softening operations, parametrize the continuous space of all softening operations. We study meta-learning a parametric S-shape curve as well as an MLP over a set of inverse rendering tasks, so that it generalizes to new and unseen differentiable rendering tasks with optimal softness.
Separating Overlapping Tissue Layers from Microscopy Images
May 22, 2019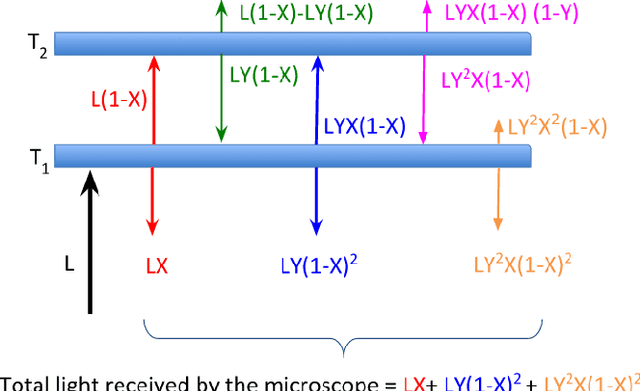


Abstract:Manual preparation of tissue slices for microscopy imaging can introduce tissue tears and overlaps. Typically, further digital processing algorithms such as registration and 3D reconstruction from tissue image stacks cannot handle images with tissue tear/overlap artifacts, and so such images are usually discarded. In this paper, we propose an imaging model and an algorithm to digitally separate overlapping tissue data of mouse brain images into two layers. We show the correctness of our model and the algorithm by comparing our results with the ground truth.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge