Yulei Hou
Relationship Discovery for Drug Recommendation
Apr 18, 2024
Abstract:Medication recommendation systems are designed to deliver personalized drug suggestions that are closely aligned with individual patient needs. Previous studies have primarily concentrated on developing medication embeddings, achieving significant progress. Nonetheless, these approaches often fall short in accurately reflecting individual patient profiles, mainly due to challenges in distinguishing between various patient conditions and the inability to establish precise correlations between specific conditions and appropriate medications. In response to these issues, we introduce DisMed, a model that focuses on patient conditions to enhance personalization. DisMed employs causal inference to discern clear, quantifiable causal links. It then examines patient conditions in depth, recognizing and adapting to the evolving nuances of these conditions, and mapping them directly to corresponding medications. Additionally, DisMed leverages data from multiple patient visits to propose combinations of medications. Comprehensive testing on real-world datasets demonstrates that DisMed not only improves the customization of patient profiles but also surpasses leading models in both precision and safety.
Dual-Granularity Medication Recommendation Based on Causal Inference
Mar 01, 2024



Abstract:As medical demands grow and machine learning technology advances, AI-based diagnostic and treatment systems are garnering increasing attention. Medication recommendation aims to integrate patients' long-term health records with medical knowledge, recommending accuracy and safe medication combinations for specific conditions. However, most existing researches treat medication recommendation systems merely as variants of traditional recommendation systems, overlooking the heterogeneity between medications and diseases. To address this challenge, we propose DGMed, a framework for medication recommendation. DGMed utilizes causal inference to uncover the connections among medical entities and presents an innovative feature alignment method to tackle heterogeneity issues. Specifically, this study first applies causal inference to analyze the quantified therapeutic effects of medications on specific diseases from historical records, uncovering potential links between medical entities. Subsequently, we integrate molecular-level knowledge, aligning the embeddings of medications and diseases within the molecular space to effectively tackle their heterogeneity. Ultimately, based on relationships at the entity level, we adaptively adjust the recommendation probabilities of medication and recommend medication combinations according to the patient's current health condition. Experimental results on a real-world dataset show that our method surpasses existing state-of-the-art baselines in four evaluation metrics, demonstrating superior performance in both accuracy and safety aspects. Compared to the sub-optimal model, our approach improved accuracy by 4.40%, reduced the risk of side effects by 6.14%, and increased time efficiency by 47.15%.
StratMed: Relevance Stratification for Low-resource Medication Recommendation
Sep 06, 2023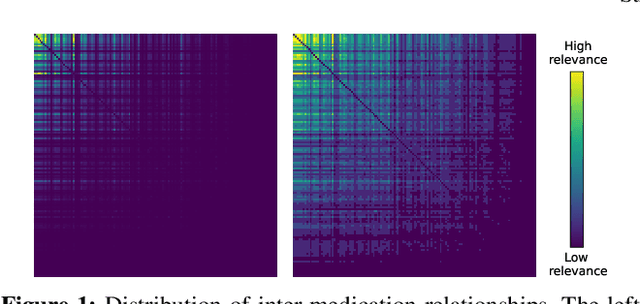
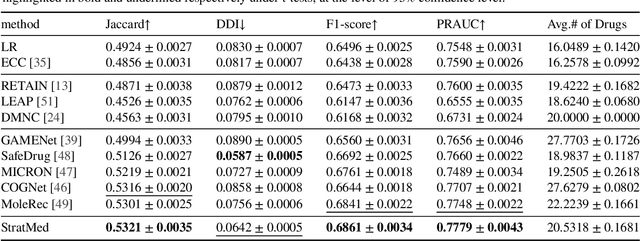
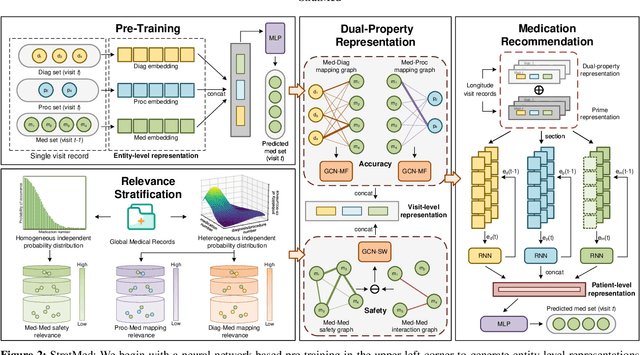
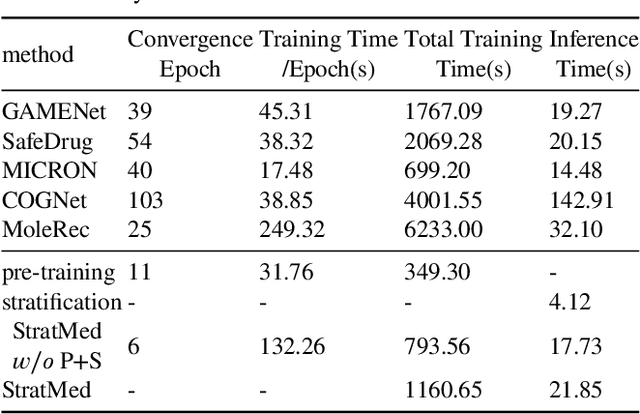
Abstract:With the growing imbalance between limited medical resources and escalating demands, AI-based clinical tasks have become paramount. Medication recommendation, as a sub-domain, aims to amalgamate longitudinal patient history with medical knowledge, assisting physicians in prescribing safer and more accurate medication combinations. Existing methods overlook the inherent long-tail distribution in medical data, lacking balanced representation between head and tail data, which leads to sub-optimal model performance. To address this challenge, we introduce StratMed, a model that incorporates an innovative relevance stratification mechanism. It harmonizes discrepancies in data long-tail distribution and strikes a balance between the safety and accuracy of medication combinations. Specifically, we first construct a pre-training method using deep learning networks to obtain entity representation. After that, we design a pyramid-like data stratification method to obtain more generalized entity relationships by reinforcing the features of unpopular entities. Based on this relationship, we designed two graph structures to express medication precision and safety at the same level to obtain visit representations. Finally, the patient's historical clinical information is fitted to generate medication combinations for the current health condition. Experiments on the MIMIC-III dataset demonstrate that our method has outperformed current state-of-the-art methods in four evaluation metrics (including safety and accuracy).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge