Yukun Guo
Multipolar Acoustic Source Reconstruction from Sparse Far-Field Data using ALOHA
Mar 12, 2023Abstract:The reconstruction of multipolar acoustic or electromagnetic sources from their far-field radiation patterns plays a crucial role in numerous practical applications. Most of the existing techniques for source reconstruction require dense multi-frequency data at the Nyquist sampling rate. Accessibility of only sparse data at a sub-sampled grid contributes to the null space of the inverse source-to-data operator, which causes significant imaging artifacts. For this purpose, additional knowledge about the source or regularization is required. In this article, we propose a novel two-stage strategy for multipolar source reconstruction from the sub-sampled sparse data that takes advantage of the sparsity of the sources in the physical domain. The data at the Nyquist sampling rate is recovered from sub-sampled data and then a conventional inversion algorithm is used to reconstruct sources. The data recovery problem is linked to a spectrum recovery problem for the signal with the finite rate of innovations (FIR) that is solved using an annihilating filter-based structured Hankel matrix completion approach (ALOHA). For an accurate reconstruction of multipolar sources, we employ a Fourier inversion algorithm. The suitability of the suggested approach for both noisy and noise-free measurements is supported by numerical evidence.
Interpretable Diabetic Retinopathy Diagnosis based on Biomarker Activation Map
Dec 13, 2022



Abstract:Deep learning classifiers provide the most accurate means of automatically diagnosing diabetic retinopathy (DR) based on optical coherence tomography (OCT) and its angiography (OCTA). The power of these models is attributable in part to the inclusion of hidden layers that provide the complexity required to achieve a desired task. However, hidden layers also render algorithm outputs difficult to interpret. Here we introduce a novel biomarker activation map (BAM) framework based on generative adversarial learning that allows clinicians to verify and understand classifiers decision-making. A data set including 456 macular scans were graded as non-referable or referable DR based on current clinical standards. A DR classifier that was used to evaluate our BAM was first trained based on this data set. The BAM generation framework was designed by combing two U-shaped generators to provide meaningful interpretability to this classifier. The main generator was trained to take referable scans as input and produce an output that would be classified by the classifier as non-referable. The BAM is then constructed as the difference image between the output and input of the main generator. To ensure that the BAM only highlights classifier-utilized biomarkers an assistant generator was trained to do the opposite, producing scans that would be classified as referable by the classifier from non-referable scans. The generated BAMs highlighted known pathologic features including nonperfusion area and retinal fluid. A fully interpretable classifier based on these highlights could help clinicians better utilize and verify automated DR diagnosis.
Automated segmentation of retinal fluid volumes from structural and angiographic optical coherence tomography using deep learning
Jun 03, 2020



Abstract:Purpose: We proposed a deep convolutional neural network (CNN), named Retinal Fluid Segmentation Network (ReF-Net) to segment volumetric retinal fluid on optical coherence tomography (OCT) volume. Methods: 3 x 3-mm OCT scans were acquired on one eye by a 70-kHz OCT commercial AngioVue system (RTVue-XR; Optovue, Inc.) from 51 participants in a clinical diabetic retinopathy (DR) study (45 with retinal edema and 6 healthy controls). A CNN with U-Net-like architecture was constructed to detect and segment the retinal fluid. Cross-sectional OCT and angiography (OCTA) scans were used for training and testing ReF-Net. The effect of including OCTA data for retinal fluid segmentation was investigated in this study. Volumetric retinal fluid can be constructed using the output of ReF-Net. Area-under-Receiver-Operating-Characteristic-curve (AROC), intersection-over-union (IoU), and F1-score were calculated to evaluate the performance of ReF-Net. Results: ReF-Net shows high accuracy (F1 = 0.864 +/- 0.084) in retinal fluid segmentation. The performance can be further improved (F1 = 0.892 +/- 0.038) by including information from both OCTA and structural OCT. ReF-Net also shows strong robustness to shadow artifacts. Volumetric retinal fluid can provide more comprehensive information than the 2D area, whether cross-sectional or en face projections. Conclusions: A deep-learning-based method can accurately segment retinal fluid volumetrically on OCT/OCTA scans with strong robustness to shadow artifacts. OCTA data can improve retinal fluid segmentation. Volumetric representations of retinal fluid are superior to 2D projections. Translational Relevance: Using a deep learning method to segment retinal fluid volumetrically has the potential to improve the diagnostic accuracy of diabetic macular edema by OCT systems.
Reconstruction of high-resolution 6x6-mm OCT angiograms using deep learning
Apr 19, 2020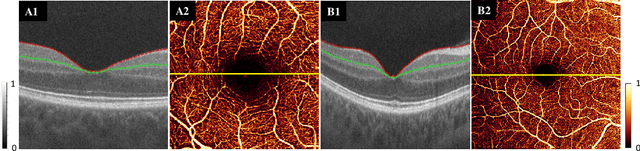
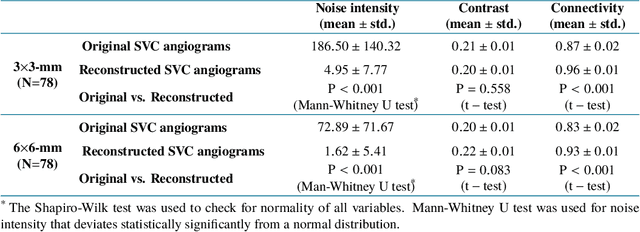
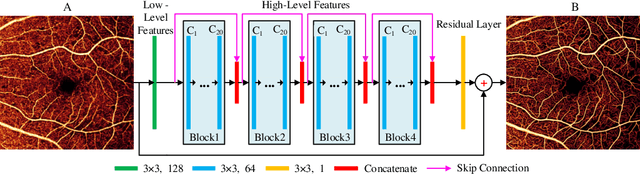
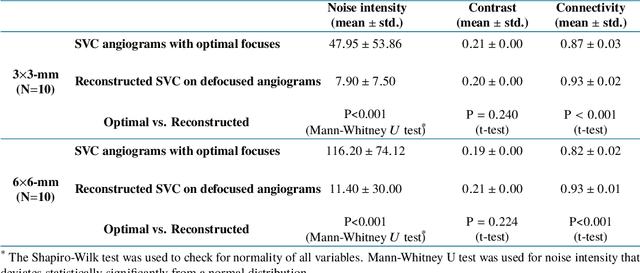
Abstract:Typical optical coherence tomographic angiography (OCTA) acquisition areas on commercial devices are 3x3- or 6x6-mm. Compared to 3x3-mm angiograms with proper sampling density, 6x6-mm angiograms have significantly lower scan quality, with reduced signal-to-noise ratio and worse shadow artifacts due to undersampling. Here, we propose a deep-learning-based high-resolution angiogram reconstruction network (HARNet) to generate enhanced 6x6-mm superficial vascular complex (SVC) angiograms. The network was trained on data from 3x3-mm and 6x6-mm angiograms from the same eyes. The reconstructed 6x6-mm angiograms have significantly lower noise intensity and better vascular connectivity than the original images. The algorithm did not generate false flow signal at the noise level presented by the original angiograms. The image enhancement produced by our algorithm may improve biomarker measurements and qualitative clinical assessment of 6x6-mm OCTA.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge