Yueh Lee
On a Hidden Property in Computational Imaging
Oct 11, 2024Abstract:Computational imaging plays a vital role in various scientific and medical applications, such as Full Waveform Inversion (FWI), Computed Tomography (CT), and Electromagnetic (EM) inversion. These methods address inverse problems by reconstructing physical properties (e.g., the acoustic velocity map in FWI) from measurement data (e.g., seismic waveform data in FWI), where both modalities are governed by complex mathematical equations. In this paper, we empirically demonstrate that despite their differing governing equations, three inverse problems (FWI, CT, and EM inversion) share a hidden property within their latent spaces. Specifically, using FWI as an example, we show that both modalities (the velocity map and seismic waveform data) follow the same set of one-way wave equations in the latent space, yet have distinct initial conditions that are linearly correlated. This suggests that after projection into the latent embedding space, the two modalities correspond to different solutions of the same equation, connected through their initial conditions. Our experiments confirm that this hidden property is consistent across all three imaging problems, providing a novel perspective for understanding these computational imaging tasks.
Deep Decomposition for Stochastic Normal-Abnormal Transport
Nov 29, 2021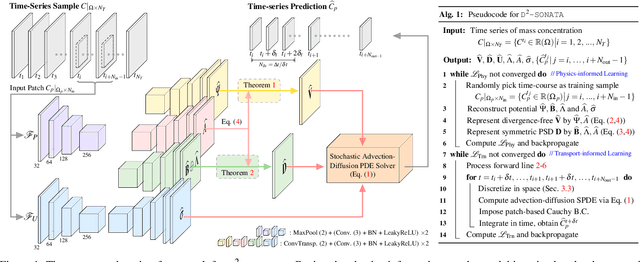
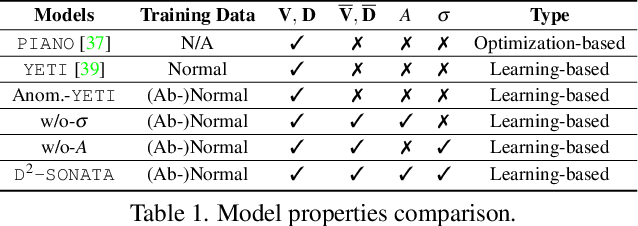
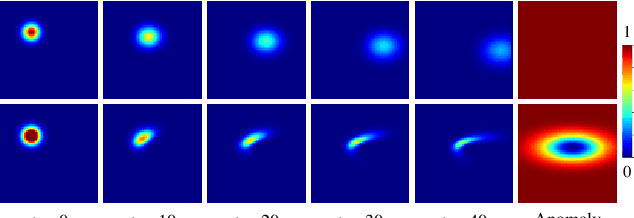
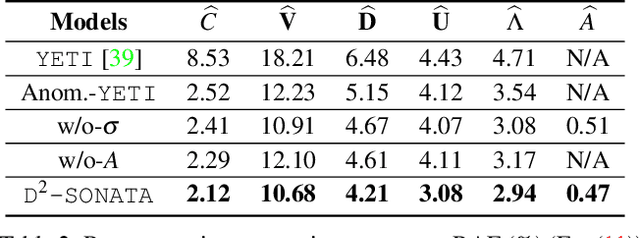
Abstract:Advection-diffusion equations describe a large family of natural transport processes, e.g., fluid flow, heat transfer, and wind transport. They are also used for optical flow and perfusion imaging computations. We develop a machine learning model, D^2-SONATA, built upon a stochastic advection-diffusion equation, which predicts the velocity and diffusion fields that drive 2D/3D image time-series of transport. In particular, our proposed model incorporates a model of transport atypicality, which isolates abnormal differences between expected normal transport behavior and the observed transport. In a medical context such a normal-abnormal decomposition can be used, for example, to quantify pathologies. Specifically, our model identifies the advection and diffusion contributions from the transport time-series and simultaneously predicts an anomaly value field to provide a decomposition into normal and abnormal advection and diffusion behavior. To achieve improved estimation performance for the velocity and diffusion-tensor fields underlying the advection-diffusion process and for the estimation of the anomaly fields, we create a 2D/3D anomaly-encoded advection-diffusion simulator, which allows for supervised learning. We further apply our model on a brain perfusion dataset from ischemic stroke patients via transfer learning. Extensive comparisons demonstrate that our model successfully distinguishes stroke lesions (abnormal) from normal brain regions, while reconstructing the underlying velocity and diffusion tensor fields.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge