Yubai Yuan
Distributional Treatment Effect Estimation across Heterogeneous Sites via Optimal Transport
Nov 12, 2025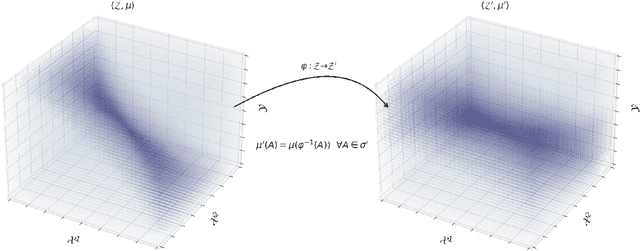
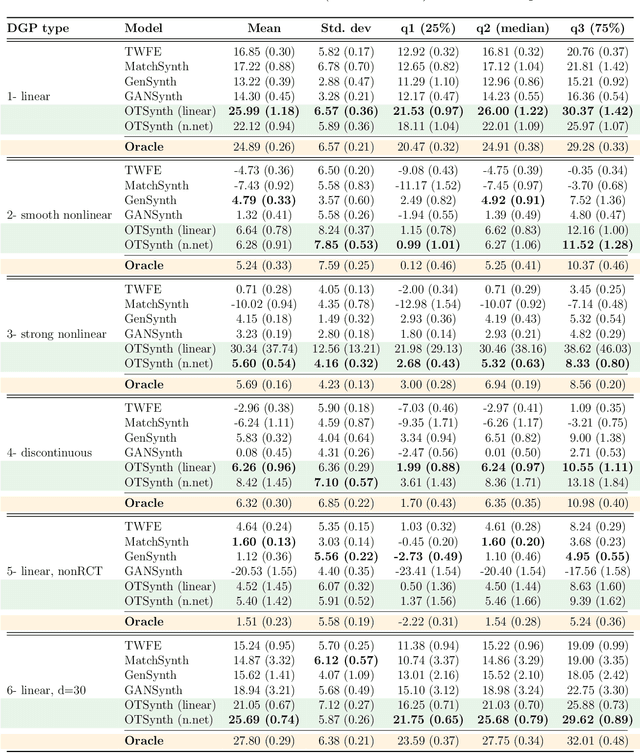
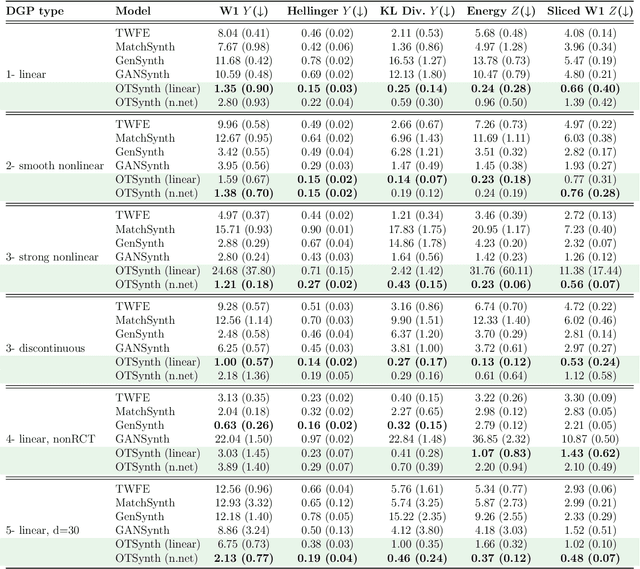
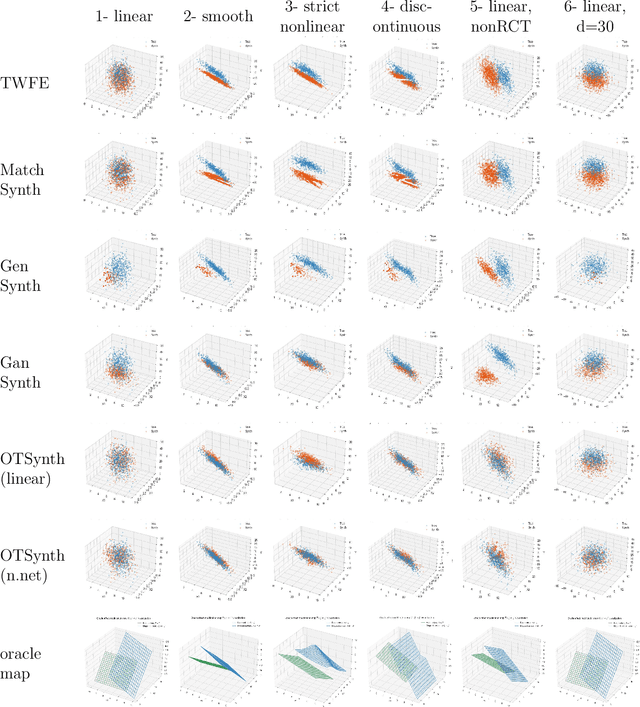
Abstract:We propose a novel framework for synthesizing counterfactual treatment group data in a target site by integrating full treatment and control group data from a source site with control group data from the target. Departing from conventional average treatment effect estimation, our approach adopts a distributional causal inference perspective by modeling treatment and control as distinct probability measures on the source and target sites. We formalize the cross-site heterogeneity (effect modification) as a push-forward transformation that maps the joint feature-outcome distribution from the source to the target site. This transformation is learned by aligning the control group distributions between sites using an Optimal Transport-based procedure, and subsequently applied to the source treatment group to generate the synthetic target treatment distribution. Under general regularity conditions, we establish theoretical guarantees for the consistency and asymptotic convergence of the synthetic treatment group data to the true target distribution. Simulation studies across multiple data-generating scenarios and a real-world application to patient-derived xenograft data demonstrate that our framework robustly recovers the full distributional properties of treatment effects.
Inferring Diffusion Structures of Heterogeneous Network Cascade
Jun 23, 2025Abstract:Network cascade refers to diffusion processes in which outcome changes within part of an interconnected population trigger a sequence of changes across the entire network. These cascades are governed by underlying diffusion networks, which are often latent. Inferring such networks is critical for understanding cascade pathways, uncovering Granger causality of interaction mechanisms among individuals, and enabling tasks such as forecasting or maximizing information propagation. In this project, we propose a novel double mixture directed graph model for inferring multi-layer diffusion networks from cascade data. The proposed model represents cascade pathways as a mixture of diffusion networks across different layers, effectively capturing the strong heterogeneity present in real-world cascades. Additionally, the model imposes layer-specific structural constraints, enabling diffusion networks at different layers to capture complementary cascading patterns at the population level. A key advantage of our model is its convex formulation, which allows us to establish both statistical and computational guarantees for the resulting diffusion network estimates. We conduct extensive simulation studies to demonstrate the model's performance in recovering diverse diffusion structures. Finally, we apply the proposed method to analyze cascades of research topics in the social sciences across U.S. universities, revealing the underlying diffusion networks of research topic propagation among institutions.
Robust Offline Active Learning on Graphs
Aug 15, 2024Abstract:We consider the problem of active learning on graphs, which has crucial applications in many real-world networks where labeling node responses is expensive. In this paper, we propose an offline active learning method that selects nodes to query by explicitly incorporating information from both the network structure and node covariates. Building on graph signal recovery theories and the random spectral sparsification technique, the proposed method adopts a two-stage biased sampling strategy that takes both informativeness and representativeness into consideration for node querying. Informativeness refers to the complexity of graph signals that are learnable from the responses of queried nodes, while representativeness refers to the capacity of queried nodes to control generalization errors given noisy node-level information. We establish a theoretical relationship between generalization error and the number of nodes selected by the proposed method. Our theoretical results demonstrate the trade-off between informativeness and representativeness in active learning. Extensive numerical experiments show that the proposed method is competitive with existing graph-based active learning methods, especially when node covariates and responses contain noises. Additionally, the proposed method is applicable to both regression and classification tasks on graphs.
Optimal Transport for Latent Integration with An Application to Heterogeneous Neuronal Activity Data
Jun 27, 2024



Abstract:Detecting dynamic patterns of task-specific responses shared across heterogeneous datasets is an essential and challenging problem in many scientific applications in medical science and neuroscience. In our motivating example of rodent electrophysiological data, identifying the dynamical patterns in neuronal activity associated with ongoing cognitive demands and behavior is key to uncovering the neural mechanisms of memory. One of the greatest challenges in investigating a cross-subject biological process is that the systematic heterogeneity across individuals could significantly undermine the power of existing machine learning methods to identify the underlying biological dynamics. In addition, many technically challenging neurobiological experiments are conducted on only a handful of subjects where rich longitudinal data are available for each subject. The low sample sizes of such experiments could further reduce the power to detect common dynamic patterns among subjects. In this paper, we propose a novel heterogeneous data integration framework based on optimal transport to extract shared patterns in complex biological processes. The key advantages of the proposed method are that it can increase discriminating power in identifying common patterns by reducing heterogeneity unrelated to the signal by aligning the extracted latent spatiotemporal information across subjects. Our approach is effective even with a small number of subjects, and does not require auxiliary matching information for the alignment. In particular, our method can align longitudinal data across heterogeneous subjects in a common latent space to capture the dynamics of shared patterns while utilizing temporal dependency within subjects.
Query-augmented Active Metric Learning
Nov 08, 2021
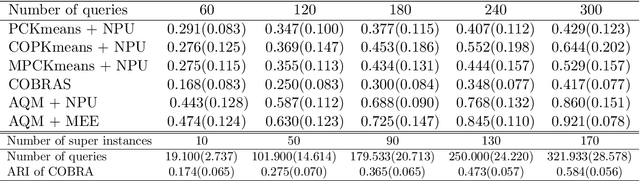
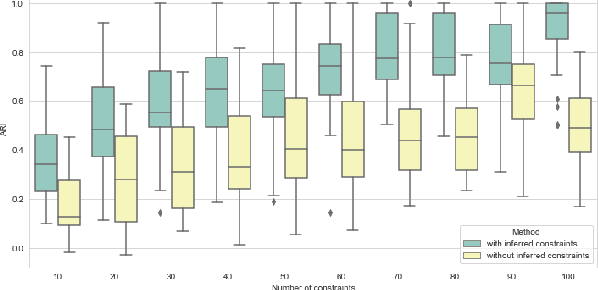
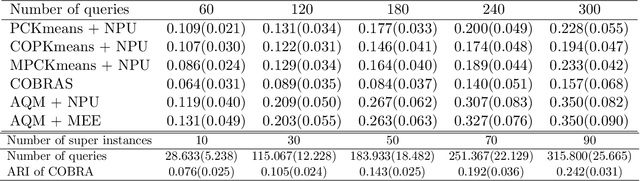
Abstract:In this paper we propose an active metric learning method for clustering with pairwise constraints. The proposed method actively queries the label of informative instance pairs, while estimating underlying metrics by incorporating unlabeled instance pairs, which leads to a more accurate and efficient clustering process. In particular, we augment the queried constraints by generating more pairwise labels to provide additional information in learning a metric to enhance clustering performance. Furthermore, we increase the robustness of metric learning by updating the learned metric sequentially and penalizing the irrelevant features adaptively. In addition, we propose a novel active query strategy that evaluates the information gain of instance pairs more accurately by incorporating the neighborhood structure, which improves clustering efficiency without extra labeling cost. In theory, we provide a tighter error bound of the proposed metric learning method utilizing augmented queries compared with methods using existing constraints only. Furthermore, we also investigate the improvement using the active query strategy instead of random selection. Numerical studies on simulation settings and real datasets indicate that the proposed method is especially advantageous when the signal-to-noise ratio between significant features and irrelevant features is low.
High-order joint embedding for multi-level link prediction
Nov 07, 2021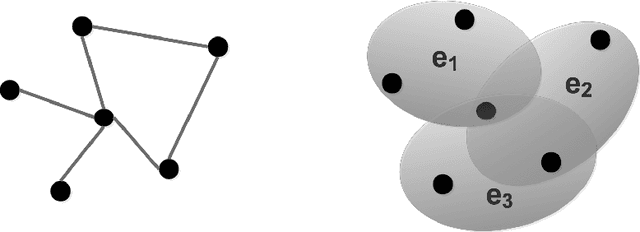
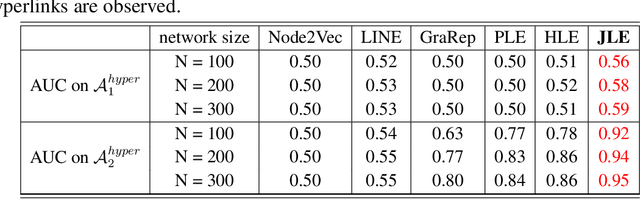
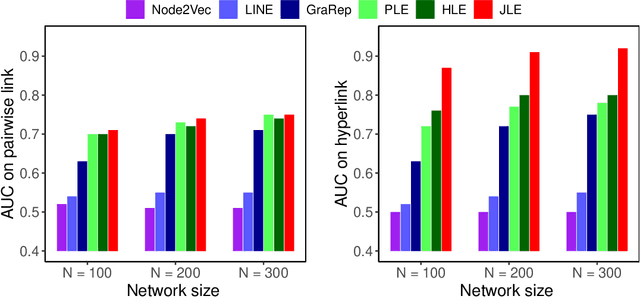
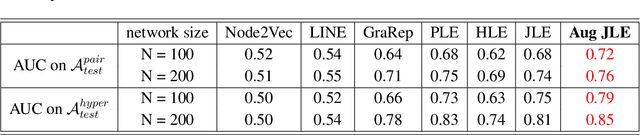
Abstract:Link prediction infers potential links from observed networks, and is one of the essential problems in network analyses. In contrast to traditional graph representation modeling which only predicts two-way pairwise relations, we propose a novel tensor-based joint network embedding approach on simultaneously encoding pairwise links and hyperlinks onto a latent space, which captures the dependency between pairwise and multi-way links in inferring potential unobserved hyperlinks. The major advantage of the proposed embedding procedure is that it incorporates both the pairwise relationships and subgroup-wise structure among nodes to capture richer network information. In addition, the proposed method introduces a hierarchical dependency among links to infer potential hyperlinks, and leads to better link prediction. In theory we establish the estimation consistency for the proposed embedding approach, and provide a faster convergence rate compared to link prediction utilizing pairwise links or hyperlinks only. Numerical studies on both simulation settings and Facebook ego-networks indicate that the proposed method improves both hyperlink and pairwise link prediction accuracy compared to existing link prediction algorithms.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge