Yuansan Liu
Local Intrinsic Dimensionality of Ground Motion Data for Early Detection of Complex Catastrophic Slope Failure
Jan 07, 2026Abstract:Local Intrinsic Dimensionality (LID) has shown strong potential for identifying anomalies and outliers in high-dimensional data across a wide range of real-world applications, including landslide failure detection in granular media. Early and accurate identification of failure zones in landslide-prone areas is crucial for effective geohazard mitigation. While existing approaches typically rely on surface displacement data analyzed through statistical or machine learning techniques, they often fall short in capturing both the spatial correlations and temporal dynamics that are inherent in such data. To address this gap, we focus on ground-monitored landslides and introduce a novel approach that jointly incorporates spatial and temporal information, enabling the detection of complex landslides and including multiple successive failures occurring in distinct areas of the same slope. To be specific, our method builds upon an existing LID-based technique, known as sLID. We extend its capabilities in three key ways. (1) Kinematic enhancement: we incorporate velocity into the sLID computation to better capture short-term temporal dependencies and deformation rate relationships. (2) Spatial fusion: we apply Bayesian estimation to aggregate sLID values across spatial neighborhoods, effectively embedding spatial correlations into the LID scores. (3) Temporal modeling: we introduce a temporal variant, tLID, that learns long-term dynamics from time series data, providing a robust temporal representation of displacement behavior. Finally, we integrate both components into a unified framework, referred to as spatiotemporal LID (stLID), to identify samples that are anomalous in either or both dimensions. Extensive experiments show that stLID consistently outperforms existing methods in failure detection precision and lead-time.
Stochastic Diffusion: A Diffusion Probabilistic Model for Stochastic Time Series Forecasting
Jun 05, 2024Abstract:Recent innovations in diffusion probabilistic models have paved the way for significant progress in image, text and audio generation, leading to their applications in generative time series forecasting. However, leveraging such abilities to model highly stochastic time series data remains a challenge. In this paper, we propose a novel Stochastic Diffusion (StochDiff) model which learns data-driven prior knowledge at each time step by utilizing the representational power of the stochastic latent spaces to model the variability of the multivariate time series data. The learnt prior knowledge helps the model to capture complex temporal dynamics and the inherent uncertainty of the data. This improves its ability to model highly stochastic time series data. Through extensive experiments on real-world datasets, we demonstrate the effectiveness of our proposed model on stochastic time series forecasting. Additionally, we showcase an application of our model for real-world surgical guidance, highlighting its potential to benefit the medical community.
Bidirectional Adversarial Autoencoders for the design of Plasmonic Metasurfaces
May 07, 2024



Abstract:Deep Learning has been a critical part of designing inverse design methods that are computationally efficient and accurate. An example of this is the design of photonic metasurfaces by using their photoluminescent spectrum as the input data to predict their topology. One fundamental challenge of these systems is their ability to represent nonlinear relationships between sets of data that have different dimensionalities. Existing design methods often implement a conditional Generative Adversarial Network in order to solve this problem, but in many cases the solution is unable to generate structures that provide multiple peaks when validated. It is demonstrated that in response to the target spectrum, the Bidirectional Adversarial Autoencoder is able to generate structures that provide multiple peaks on several occasions. As a result the proposed model represents an important advance towards the generation of nonlinear photonic metasurfaces that can be used in advanced metasurface design.
Time Series Representation Learning with Supervised Contrastive Temporal Transformer
Mar 16, 2024Abstract:Finding effective representations for time series data is a useful but challenging task. Several works utilize self-supervised or unsupervised learning methods to address this. However, there still remains the open question of how to leverage available label information for better representations. To answer this question, we exploit pre-existing techniques in time series and representation learning domains and develop a simple, yet novel fusion model, called: \textbf{S}upervised \textbf{CO}ntrastive \textbf{T}emporal \textbf{T}ransformer (SCOTT). We first investigate suitable augmentation methods for various types of time series data to assist with learning change-invariant representations. Secondly, we combine Transformer and Temporal Convolutional Networks in a simple way to efficiently learn both global and local features. Finally, we simplify Supervised Contrastive Loss for representation learning of labelled time series data. We preliminarily evaluate SCOTT on a downstream task, Time Series Classification, using 45 datasets from the UCR archive. The results show that with the representations learnt by SCOTT, even a weak classifier can perform similar to or better than existing state-of-the-art models (best performance on 23/45 datasets and highest rank against 9 baseline models). Afterwards, we investigate SCOTT's ability to address a real-world task, online Change Point Detection (CPD), on two datasets: a human activity dataset and a surgical patient dataset. We show that the model performs with high reliability and efficiency on the online CPD problem ($\sim$98\% and $\sim$97\% area under precision-recall curve respectively). Furthermore, we demonstrate the model's potential in tackling early detection and show it performs best compared to other candidates.
Time-Transformer: Integrating Local and Global Features for Better Time Series Generation
Dec 18, 2023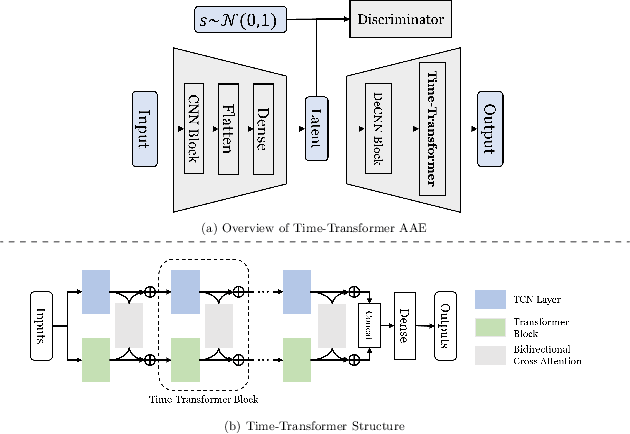

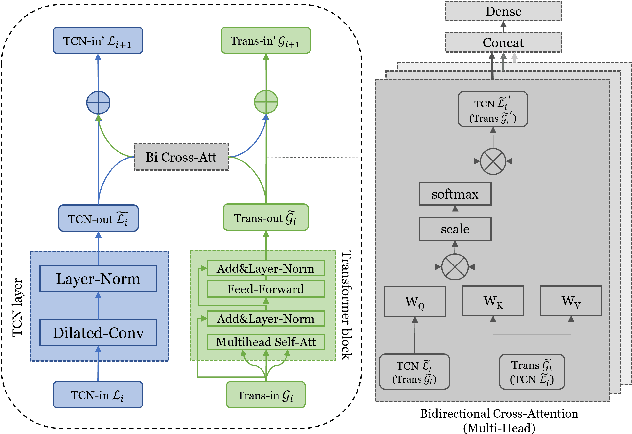

Abstract:Generating time series data is a promising approach to address data deficiency problems. However, it is also challenging due to the complex temporal properties of time series data, including local correlations as well as global dependencies. Most existing generative models have failed to effectively learn both the local and global properties of time series data. To address this open problem, we propose a novel time series generative model named 'Time-Transformer AAE', which consists of an adversarial autoencoder (AAE) and a newly designed architecture named 'Time-Transformer' within the decoder. The Time-Transformer first simultaneously learns local and global features in a layer-wise parallel design, combining the abilities of Temporal Convolutional Networks and Transformer in extracting local features and global dependencies respectively. Second, a bidirectional cross attention is proposed to provide complementary guidance across the two branches and achieve proper fusion between local and global features. Experimental results demonstrate that our model can outperform existing state-of-the-art models in 5 out of 6 datasets, specifically on those with data containing both global and local properties. Furthermore, we highlight our model's advantage on handling this kind of data via an artificial dataset. Finally, we show our model's ability to address a real-world problem: data augmentation to support learning with small datasets and imbalanced datasets.
De Novo Molecular Generation with Stacked Adversarial Model
Oct 24, 2021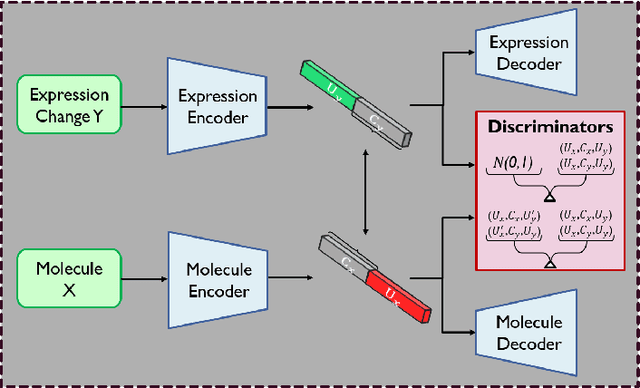

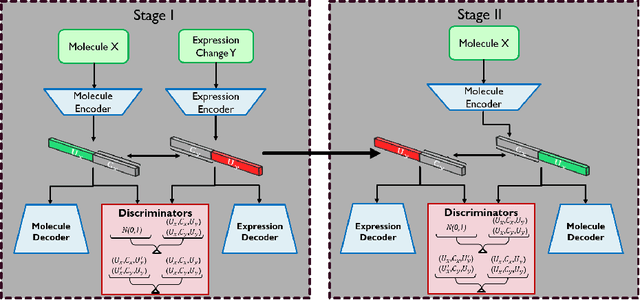

Abstract:Generating novel drug molecules with desired biological properties is a time consuming and complex task. Conditional generative adversarial models have recently been proposed as promising approaches for de novo drug design. In this paper, we propose a new generative model which extends an existing adversarial autoencoder (AAE) based model by stacking two models together. Our stacked approach generates more valid molecules, as well as molecules that are more similar to known drugs. We break down this challenging task into two sub-problems. A first stage model to learn primitive features from the molecules and gene expression data. A second stage model then takes these features to learn properties of the molecules and refine more valid molecules. Experiments and comparison to baseline methods on the LINCS L1000 dataset demonstrate that our proposed model has promising performance for molecular generation.
Pandemic model with data-driven phase detection, a study using COVID-19 data
Oct 24, 2021
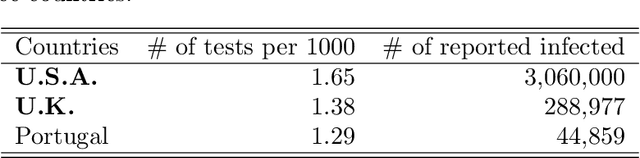
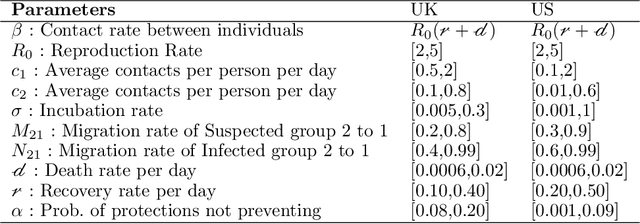

Abstract:The recent COVID-19 pandemic has promoted vigorous scientific activity in an effort to understand, advice and control the pandemic. Data is now freely available at a staggering rate worldwide. Unfortunately, this unprecedented level of information contains a variety of data sources and formats, and the models do not always conform to the description of the data. Health officials have recognized the need for more accurate models that can adjust to sudden changes, such as produced by changes in behavior or social restrictions. In this work we formulate a model that fits a ``SIR''-type model concurrently with a statistical change detection test on the data. The result is a piece wise autonomous ordinary differential equation, whose parameters change at various points in time (automatically learned from the data). The main contributions of our model are: (a) providing interpretation of the parameters, (b) determining which parameters of the model are more important to produce changes in the spread of the disease, and (c) using data-driven discovery of sudden changes in the evolution of the pandemic. Together, these characteristics provide a new model that better describes the situation and thus, provides better quality of information for decision making.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge