Yinsheng He
Enhancing Weakly-Supervised Histopathology Image Segmentation with Knowledge Distillation on MIL-Based Pseudo-Labels
Jul 14, 2024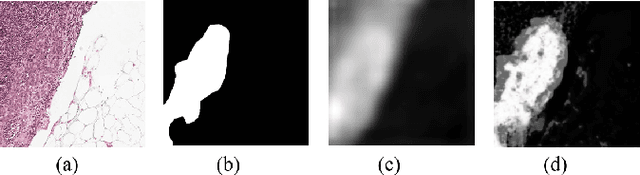

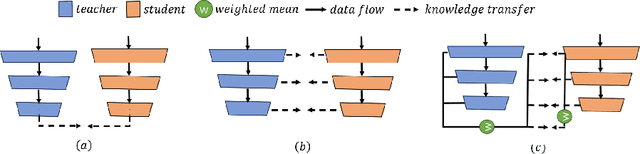

Abstract:Segmenting tumors in histological images is vital for cancer diagnosis. While fully supervised models excel with pixel-level annotations, creating such annotations is labor-intensive and costly. Accurate histopathology image segmentation under weakly-supervised conditions with coarse-grained image labels is still a challenging problem. Although multiple instance learning (MIL) has shown promise in segmentation tasks, surprisingly, no previous pseudo-supervision methods have used MIL-based outputs as pseudo-masks for training. We suspect this stems from concerns over noises in MIL results affecting pseudo supervision quality. To explore the potential of leveraging MIL-based segmentation for pseudo supervision, we propose a novel distillation framework for histopathology image segmentation. This framework introduces a iterative fusion-knowledge distillation strategy, enabling the student model to learn directly from the teacher's comprehensive outcomes. Through dynamic role reversal between the fixed teacher and learnable student models and the incorporation of weighted cross-entropy loss for model optimization, our approach prevents performance deterioration and noise amplification during knowledge distillation. Experimental results on public histopathology datasets, Camelyon16 and Digestpath2019, demonstrate that our approach not only complements various MIL-based segmentation methods but also significantly enhances their performance. Additionally, our method achieves new SOTA in the field.
BMAD: Benchmarks for Medical Anomaly Detection
Jun 28, 2023Abstract:Anomaly detection (AD) is a fundamental research problem in machine learning and computer vision, with practical applications in industrial inspection, video surveillance, and medical diagnosis. In medical imaging, AD is especially vital for detecting and diagnosing anomalies that may indicate rare diseases or conditions. However, there is a lack of a universal and fair benchmark for evaluating AD methods on medical images, which hinders the development of more generalized and robust AD methods in this specific domain. To bridge this gap, we introduce a comprehensive evaluation benchmark for assessing anomaly detection methods on medical images. This benchmark encompasses six reorganized datasets from five medical domains (i.e. brain MRI, liver CT, retinal OCT, chest X-ray, and digital histopathology) and three key evaluation metrics, and includes a total of fourteen state-of-the-art AD algorithms. This standardized and well-curated medical benchmark with the well-structured codebase enables comprehensive comparisons among recently proposed anomaly detection methods. It will facilitate the community to conduct a fair comparison and advance the field of AD on medical imaging. More information on BMAD is available in our GitHub repository: https://github.com/DorisBao/BMAD
Whole-slide-imaging Cancer Metastases Detection and Localization with Limited Tumorous Data
Mar 18, 2023



Abstract:Recently, various deep learning methods have shown significant successes in medical image analysis, especially in the detection of cancer metastases in hematoxylin and eosin (H&E) stained whole-slide images (WSIs). However, in order to obtain good performance, these research achievements rely on hundreds of well-annotated WSIs. In this study, we tackle the tumor localization and detection problem under the setting of few labeled whole slide images and introduce a patch-based analysis pipeline based on the latest reverse knowledge distillation architecture. To address the extremely unbalanced normal and tumorous samples in training sample collection, we applied the focal loss formula to the representation similarity metric for model optimization. Compared with prior arts, our method achieves similar performance by less than ten percent of training samples on the public Camelyon16 dataset. In addition, this is the first work that show the great potential of the knowledge distillation models in computational histopathology.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge