Enhancing Weakly-Supervised Histopathology Image Segmentation with Knowledge Distillation on MIL-Based Pseudo-Labels
Paper and Code
Jul 14, 2024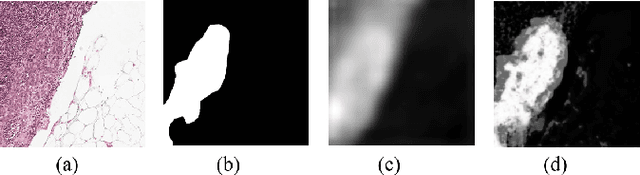

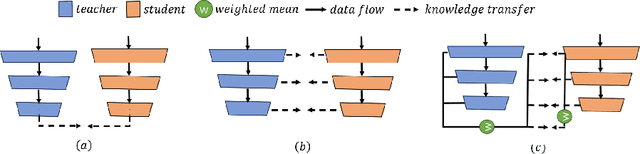

Segmenting tumors in histological images is vital for cancer diagnosis. While fully supervised models excel with pixel-level annotations, creating such annotations is labor-intensive and costly. Accurate histopathology image segmentation under weakly-supervised conditions with coarse-grained image labels is still a challenging problem. Although multiple instance learning (MIL) has shown promise in segmentation tasks, surprisingly, no previous pseudo-supervision methods have used MIL-based outputs as pseudo-masks for training. We suspect this stems from concerns over noises in MIL results affecting pseudo supervision quality. To explore the potential of leveraging MIL-based segmentation for pseudo supervision, we propose a novel distillation framework for histopathology image segmentation. This framework introduces a iterative fusion-knowledge distillation strategy, enabling the student model to learn directly from the teacher's comprehensive outcomes. Through dynamic role reversal between the fixed teacher and learnable student models and the incorporation of weighted cross-entropy loss for model optimization, our approach prevents performance deterioration and noise amplification during knowledge distillation. Experimental results on public histopathology datasets, Camelyon16 and Digestpath2019, demonstrate that our approach not only complements various MIL-based segmentation methods but also significantly enhances their performance. Additionally, our method achieves new SOTA in the field.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge