Yilai Li
CryoFM: A Flow-based Foundation Model for Cryo-EM Densities
Oct 11, 2024Abstract:Cryo-electron microscopy (cryo-EM) is a powerful technique in structural biology and drug discovery, enabling the study of biomolecules at high resolution. Significant advancements by structural biologists using cryo-EM have led to the production of over 38,626 protein density maps at various resolutions1. However, cryo-EM data processing algorithms have yet to fully benefit from our knowledge of biomolecular density maps, with only a few recent models being data-driven but limited to specific tasks. In this study, we present CryoFM, a foundation model designed as a generative model, learning the distribution of high-quality density maps and generalizing effectively to downstream tasks. Built on flow matching, CryoFM is trained to accurately capture the prior distribution of biomolecular density maps. Furthermore, we introduce a flow posterior sampling method that leverages CRYOFM as a flexible prior for several downstream tasks in cryo-EM and cryo-electron tomography (cryo-ET) without the need for fine-tuning, achieving state-of-the-art performance on most tasks and demonstrating its potential as a foundational model for broader applications in these fields.
CryoRL: Reinforcement Learning Enables Efficient Cryo-EM Data Collection
Apr 15, 2022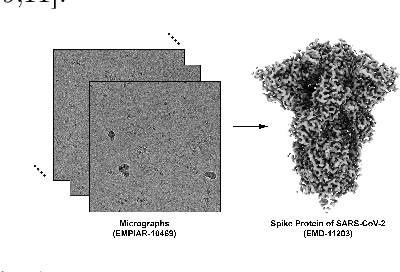

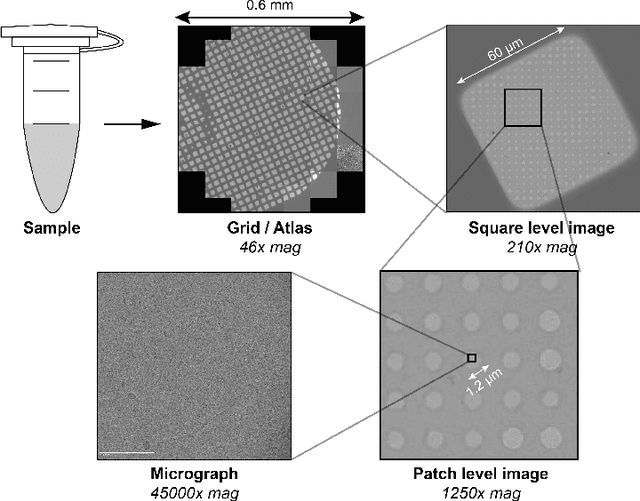
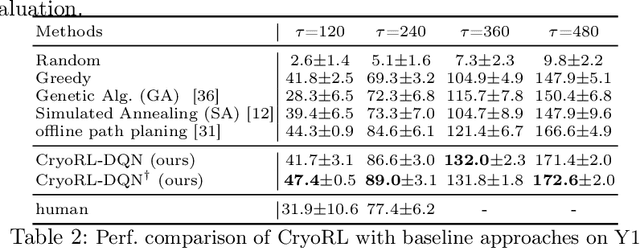
Abstract:Single-particle cryo-electron microscopy (cryo-EM) has become one of the mainstream structural biology techniques because of its ability to determine high-resolution structures of dynamic bio-molecules. However, cryo-EM data acquisition remains expensive and labor-intensive, requiring substantial expertise. Structural biologists need a more efficient and objective method to collect the best data in a limited time frame. We formulate the cryo-EM data collection task as an optimization problem in this work. The goal is to maximize the total number of good images taken within a specified period. We show that reinforcement learning offers an effective way to plan cryo-EM data collection, successfully navigating heterogenous cryo-EM grids. The approach we developed, cryoRL, demonstrates better performance than average users for data collection under similar settings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge