Yaxin Bi
Design and Development of a Robust Tolerance Optimisation Framework for Automated Optical Inspection in Semiconductor Manufacturing
May 06, 2025Abstract:Automated Optical Inspection (AOI) is widely used across various industries, including surface mount technology in semiconductor manufacturing. One of the key challenges in AOI is optimising inspection tolerances. Traditionally, this process relies heavily on the expertise and intuition of engineers, making it subjective and prone to inconsistency. To address this, we are developing an intelligent, data-driven approach to optimise inspection tolerances in a more objective and consistent manner. Most existing research in this area focuses primarily on minimising false calls, often at the risk of allowing actual defects to go undetected. This oversight can compromise product quality, especially in critical sectors such as medical, defence, and automotive industries. Our approach introduces the use of percentile rank, amongst other logical strategies, to ensure that genuine defects are not overlooked. With continued refinement, our method aims to reach a point where every flagged item is a true defect, thereby eliminating the need for manual inspection. Our proof of concept achieved an 18% reduction in false calls at the 80th percentile rank, while maintaining a 100% recall rate. This makes the system both efficient and reliable, offering significant time and cost savings.
Rethinking the Encoding of Satellite Image Time Series
May 03, 2023
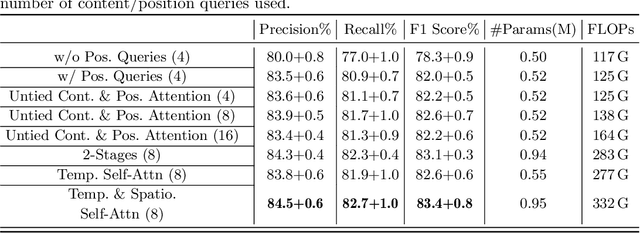

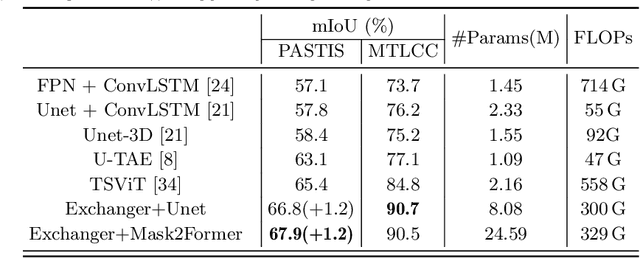
Abstract:Representation learning of Satellite Image Time Series (SITS) presents its unique challenges, such as prohibitive computation burden caused by high spatiotemporal resolutions, irregular acquisition times, and complex spatiotemporal interactions, leading to highly-specialized neural network architectures for SITS analysis. Despite the promising results achieved by some pioneering work, we argue that satisfactory representation learning paradigms have not yet been established for SITS analysis, causing an isolated island where transferring successful paradigms or the latest advances from Computer Vision (CV) to SITS is arduous. In this paper, we develop a unique perspective of SITS processing as a direct set prediction problem, inspired by the recent trend in adopting query-based transformer decoders to streamline the object detection or image segmentation pipeline, and further propose to decompose the representation learning process of SITS into three explicit steps: collect--update--distribute, which is computationally efficient and suits for irregularly-sampled and asynchronous temporal observations. Facilitated by the unique reformulation and effective feature extraction framework proposed, our models pre-trained on pixel-set format input and then fine-tuned on downstream dense prediction tasks by simply appending a commonly-used segmentation network have attained new state-of-the-art (SoTA) results on PASTIS dataset compared to bespoke neural architectures such as U-TAE. Furthermore, the clear separation, conceptually and practically, between temporal and spatial components in the panoptic segmentation pipeline of SITS allows us to leverage the recent advances in CV, such as Mask2Former, a universal segmentation architecture, resulting in a noticeable 8.8 points increase in PQ compared to the best score reported so far.
Tampered VAE for Improved Satellite Image Time Series Classification
Mar 30, 2022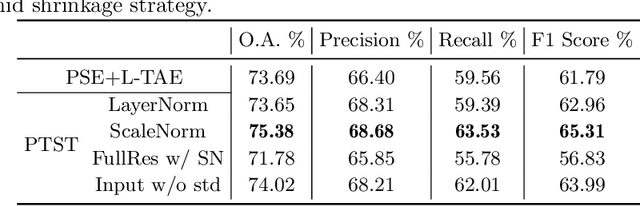
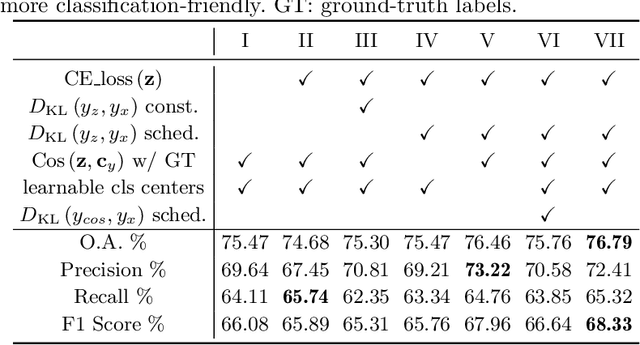
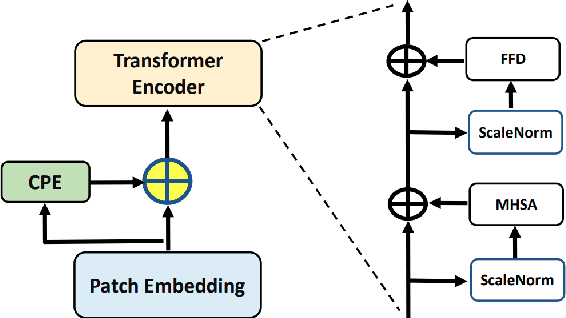
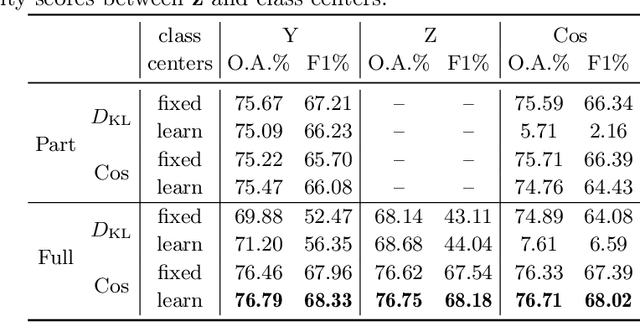
Abstract:The unprecedented availability of spatial and temporal high-resolution satellite image time series (SITS) for crop type mapping is believed to necessitate deep learning architectures to accommodate challenges arising from both dimensions. Recent state-of-the-art deep learning models have shown promising results by stacking spatial and temporal encoders. However, we present a Pyramid Time-Series Transformer (PTST) that operates solely on the temporal dimension, i.e., neglecting the spatial dimension, can produce superior results with a drastic reduction in GPU memory consumption and easy extensibility. Furthermore, we augment it to perform semi-supervised learning by proposing a classification-friendly VAE framework that introduces clustering mechanisms into latent space and can promote linear separability therein. Consequently, a few principal axes of the latent space can explain the majority of variance in raw data. Meanwhile, the VAE framework with proposed tweaks can maintain competitive classification performance as its purely discriminative counterpart when only $40\%$ of labelled data is used. We hope the proposed framework can serve as a baseline for crop classification with SITS for its modularity and simplicity.
Incremental Transductive Learning Approaches to Schistosomiasis Vector Classification
Apr 06, 2017
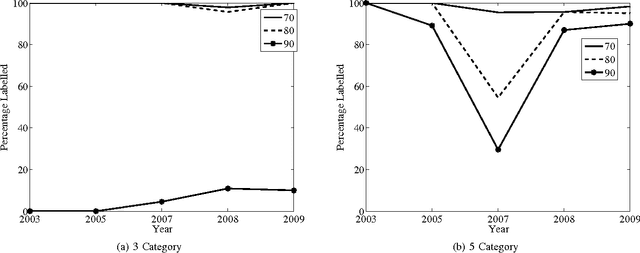

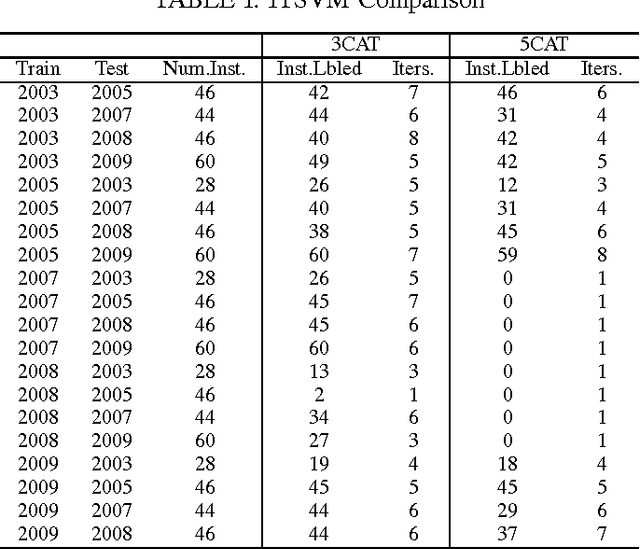
Abstract:The key issues pertaining to collection of epidemic disease data for our analysis purposes are that it is a labour intensive, time consuming and expensive process resulting in availability of sparse sample data which we use to develop prediction models. To address this sparse data issue, we present novel Incremental Transductive methods to circumvent the data collection process by applying previously acquired data to provide consistent, confidence-based labelling alternatives to field survey research. We investigated various reasoning approaches for semisupervised machine learning including Bayesian models for labelling data. The results show that using the proposed methods, we can label instances of data with a class of vector density at a high level of confidence. By applying the Liberal and Strict Training Approaches, we provide a labelling and classification alternative to standalone algorithms. The methods in this paper are components in the process of reducing the proliferation of the Schistosomiasis disease and its effects.
An Efficient Triplet-based Algorithm for Evidential Reasoning
Jun 27, 2012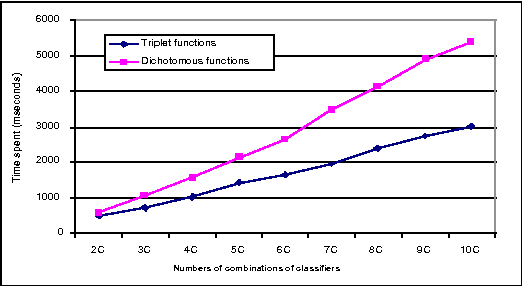
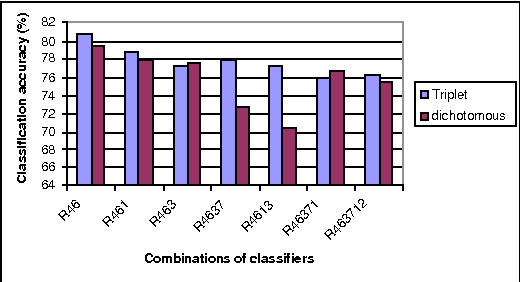
Abstract:Linear-time computational techniques have been developed for combining evidence which is available on a number of contending hypotheses. They offer a means of making the computation-intensive calculations involved more efficient in certain circumstances. Unfortunately, they restrict the orthogonal sum of evidential functions to the dichotomous structure applies only to elements and their complements. In this paper, we present a novel evidence structure in terms of a triplet and a set of algorithms for evidential reasoning. The merit of this structure is that it divides a set of evidence into three subsets, distinguishing trivial evidential elements from important ones focusing some particular elements. It avoids the deficits of the dichotomous structure in representing the preference of evidence and estimating the basic probability assignment of evidence. We have established a formalism for this structure and the general formulae for combining pieces of evidence in the form of the triplet, which have been theoretically justified.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge