Yassine El Ouahidi
Ministral 3
Jan 13, 2026Abstract:We introduce the Ministral 3 series, a family of parameter-efficient dense language models designed for compute and memory constrained applications, available in three model sizes: 3B, 8B, and 14B parameters. For each model size, we release three variants: a pretrained base model for general-purpose use, an instruction finetuned, and a reasoning model for complex problem-solving. In addition, we present our recipe to derive the Ministral 3 models through Cascade Distillation, an iterative pruning and continued training with distillation technique. Each model comes with image understanding capabilities, all under the Apache 2.0 license.
REVE: A Foundation Model for EEG -- Adapting to Any Setup with Large-Scale Pretraining on 25,000 Subjects
Oct 24, 2025Abstract:Foundation models have transformed AI by reducing reliance on task-specific data through large-scale pretraining. While successful in language and vision, their adoption in EEG has lagged due to the heterogeneity of public datasets, which are collected under varying protocols, devices, and electrode configurations. Existing EEG foundation models struggle to generalize across these variations, often restricting pretraining to a single setup, resulting in suboptimal performance, in particular under linear probing. We present REVE (Representation for EEG with Versatile Embeddings), a pretrained model explicitly designed to generalize across diverse EEG signals. REVE introduces a novel 4D positional encoding scheme that enables it to process signals of arbitrary length and electrode arrangement. Using a masked autoencoding objective, we pretrain REVE on over 60,000 hours of EEG data from 92 datasets spanning 25,000 subjects, representing the largest EEG pretraining effort to date. REVE achieves state-of-the-art results on 10 downstream EEG tasks, including motor imagery classification, seizure detection, sleep staging, cognitive load estimation, and emotion recognition. With little to no fine-tuning, it demonstrates strong generalization, and nuanced spatio-temporal modeling. We release code, pretrained weights, and tutorials to support standardized EEG research and accelerate progress in clinical neuroscience.
Unsupervised Adaptive Deep Learning Method For BCI Motor Imagery Decoding
Mar 15, 2024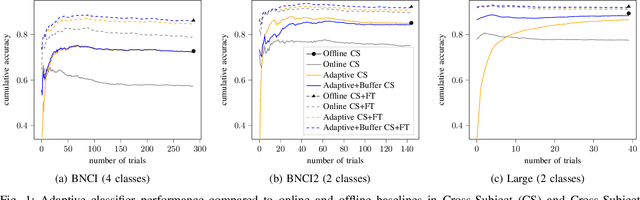
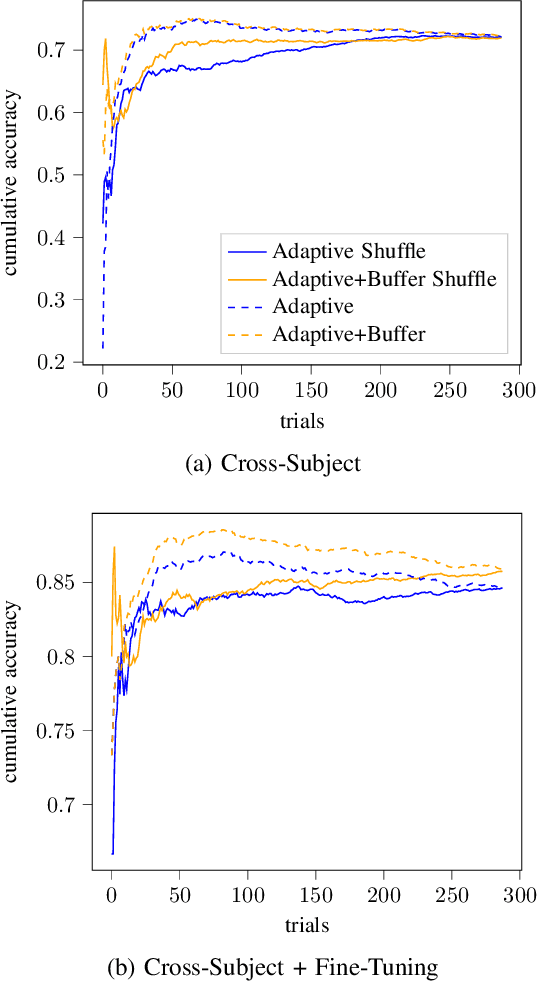
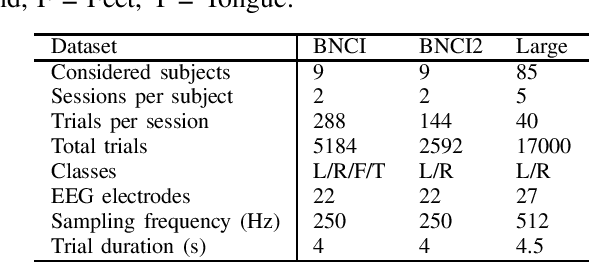
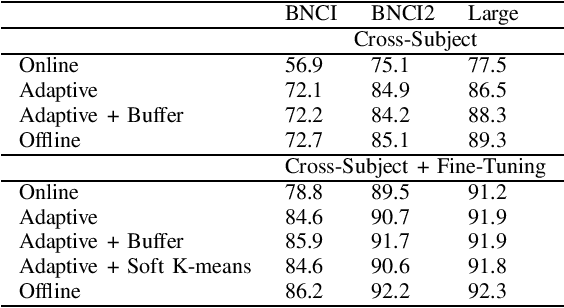
Abstract:In the context of Brain-Computer Interfaces, we propose an adaptive method that reaches offline performance level while being usable online without requiring supervision. Interestingly, our method does not require retraining the model, as it consists in using a frozen efficient deep learning backbone while continuously realigning data, both at input and latent spaces, based on streaming observations. We demonstrate its efficiency for Motor Imagery brain decoding from electroencephalography data, considering challenging cross-subject scenarios. For reproducibility, we share the code of our experiments.
A Strong and Simple Deep Learning Baseline for BCI MI Decoding
Sep 11, 2023
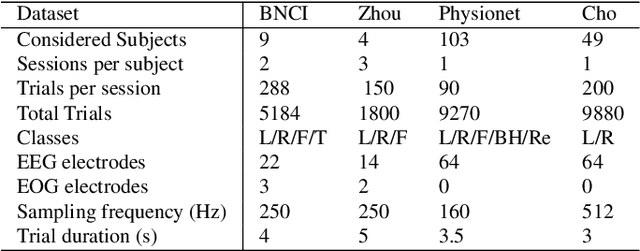


Abstract:We propose EEG-SimpleConv, a straightforward 1D convolutional neural network for Motor Imagery decoding in BCI. Our main motivation is to propose a very simple baseline to compare to, using only very standard ingredients from the literature. We evaluate its performance on four EEG Motor Imagery datasets, including simulated online setups, and compare it to recent Deep Learning and Machine Learning approaches. EEG-SimpleConv is at least as good or far more efficient than other approaches, showing strong knowledge-transfer capabilities across subjects, at the cost of a low inference time. We advocate that using off-the-shelf ingredients rather than coming with ad-hoc solutions can significantly help the adoption of Deep Learning approaches for BCI. We make the code of the models and the experiments accessible.
Spatial Graph Signal Interpolation with an Application for Merging BCI Datasets with Various Dimensionalities
Oct 28, 2022Abstract:BCI Motor Imagery datasets usually are small and have different electrodes setups. When training a Deep Neural Network, one may want to capitalize on all these datasets to increase the amount of data available and hence obtain good generalization results. To this end, we introduce a spatial graph signal interpolation technique, that allows to interpolate efficiently multiple electrodes. We conduct a set of experiments with five BCI Motor Imagery datasets comparing the proposed interpolation with spherical splines interpolation. We believe that this work provides novel ideas on how to leverage graphs to interpolate electrodes and on how to homogenize multiple datasets.
Pruning Graph Convolutional Networks to select meaningful graph frequencies for fMRI decoding
Mar 09, 2022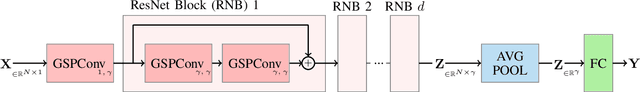
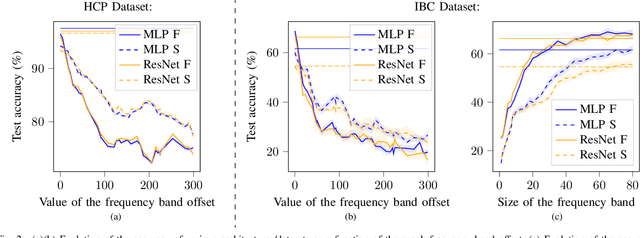
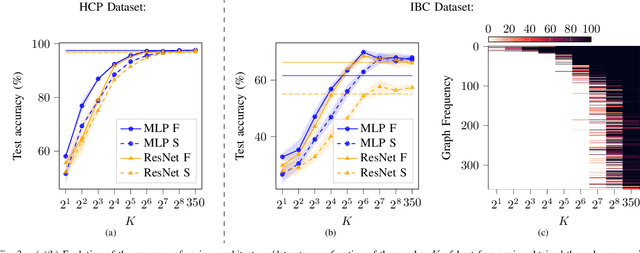
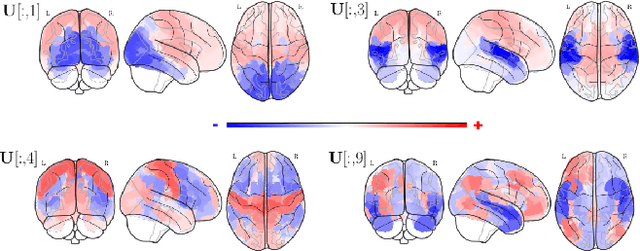
Abstract:Graph Signal Processing is a promising framework to manipulate brain signals as it allows to encompass the spatial dependencies between the activity in regions of interest in the brain. In this work, we are interested in better understanding what are the graph frequencies that are the most useful to decode fMRI signals. To this end, we introduce a deep learning architecture and adapt a pruning methodology to automatically identify such frequencies. We experiment with various datasets, architectures and graphs, and show that low graph frequencies are consistently identified as the most important for fMRI decoding, with a stronger contribution for the functional graph over the structural one. We believe that this work provides novel insights on how graph-based methods can be deployed to increase fMRI decoding accuracy and interpretability.
An Approach for Clustering Subjects According to Similarities in Cell Distributions within Biopsies
Jul 06, 2020



Abstract:In this paper, we introduce a novel and interpretable methodology to cluster subjects suffering from cancer, based on features extracted from their biopsies. Contrary to existing approaches, we propose here to capture complex patterns in the repartitions of their cells using histograms, and compare subjects on the basis of these repartitions. We describe here our complete workflow, including creation of the database, cells segmentation and phenotyping, computation of complex features, choice of a distance function between features, clustering between subjects using that distance, and survival analysis of obtained clusters. We illustrate our approach on a database of hematoxylin and eosin (H&E)-stained tissues of subjects suffering from Stage I lung adenocarcinoma, where our results match existing knowledge in prognosis estimation with high confidence.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge