Xiao Gan
Pathological Primitive Segmentation Based on Visual Foundation Model with Zero-Shot Mask Generation
Apr 12, 2024Abstract:Medical image processing usually requires a model trained with carefully crafted datasets due to unique image characteristics and domain-specific challenges, especially in pathology. Primitive detection and segmentation in digitized tissue samples are essential for objective and automated diagnosis and prognosis of cancer. SAM (Segment Anything Model) has recently been developed to segment general objects from natural images with high accuracy, but it requires human prompts to generate masks. In this work, we present a novel approach that adapts pre-trained natural image encoders of SAM for detection-based region proposals. Regions proposed by a pre-trained encoder are sent to cascaded feature propagation layers for projection. Then, local semantic and global context is aggregated from multi-scale for bounding box localization and classification. Finally, the SAM decoder uses the identified bounding boxes as essential prompts to generate a comprehensive primitive segmentation map. The entire base framework, SAM, requires no additional training or fine-tuning but could produce an end-to-end result for two fundamental segmentation tasks in pathology. Our method compares with state-of-the-art models in F1 score for nuclei detection and binary/multiclass panoptic(bPQ/mPQ) and mask quality(dice) for segmentation quality on the PanNuke dataset while offering end-to-end efficiency. Our model also achieves remarkable Average Precision (+4.5%) on the secondary dataset (HuBMAP Kidney) compared to Faster RCNN. The code is publicly available at https://github.com/learner-codec/autoprom_sam.
Network Medicine Framework for Identifying Drug Repurposing Opportunities for COVID-19
Apr 15, 2020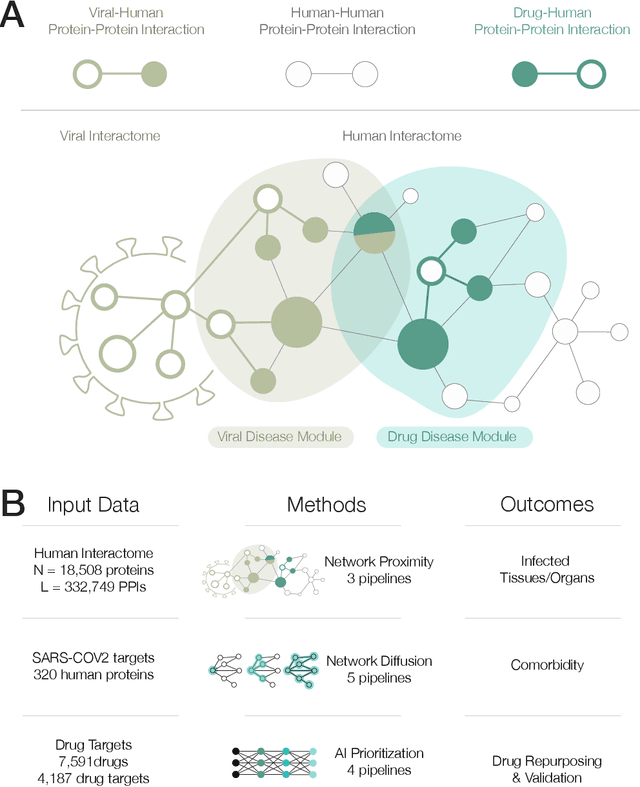
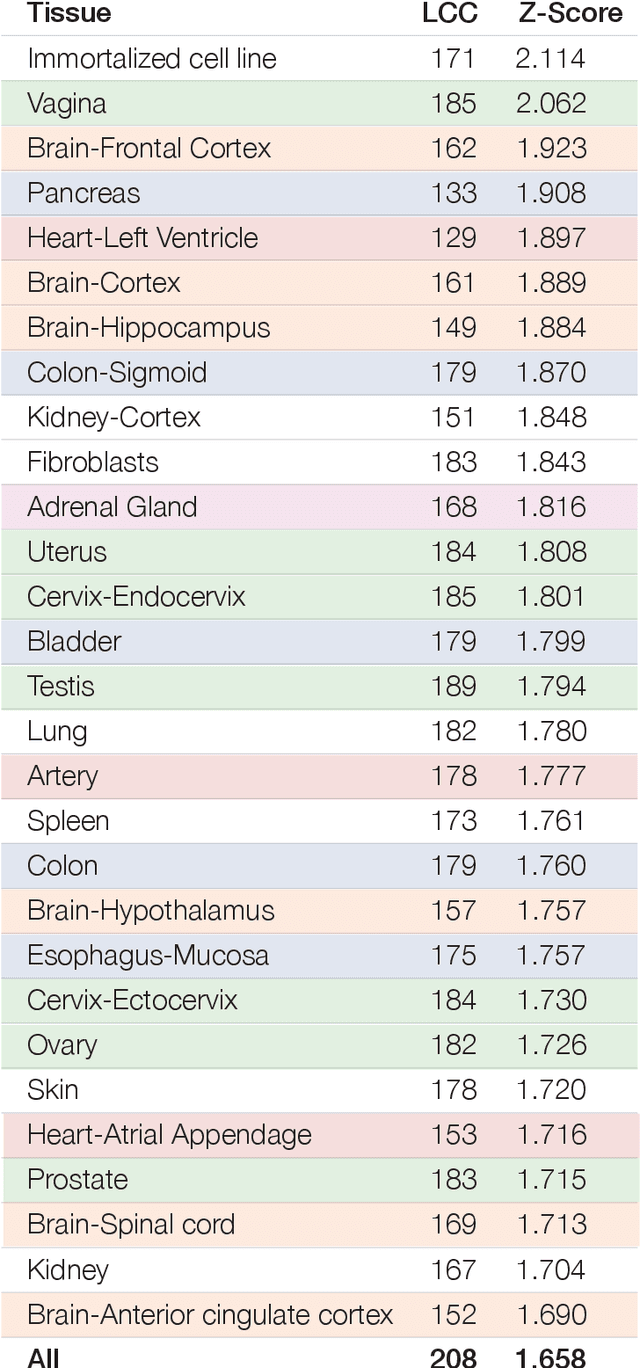
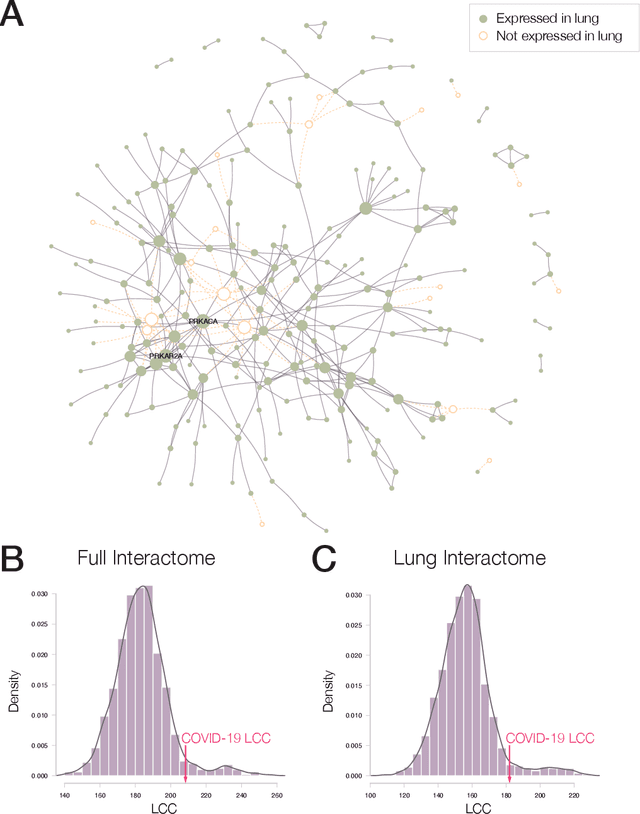
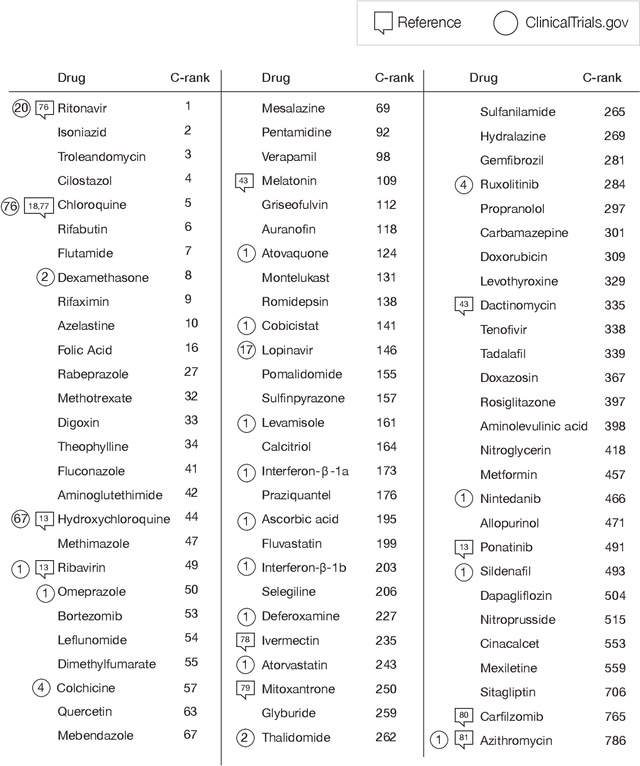
Abstract:The COVID-19 pandemic demands the rapid identification of drug-repurpusing candidates. In the past decade, network medicine had developed a framework consisting of a series of quantitative approaches and predictive tools to study host-pathogen interactions, unveil the molecular mechanisms of the infection, identify comorbidities as well as rapidly detect drug repurpusing candidates. Here, we adapt the network-based toolset to COVID-19, recovering the primary pulmonary manifestations of the virus in the lung as well as observed comorbidities associated with cardiovascular diseases. We predict that the virus can manifest itself in other tissues, such as the reproductive system, and brain regions, moreover we predict neurological comorbidities. We build on these findings to deploy three network-based drug repurposing strategies, relying on network proximity, diffusion, and AI-based metrics, allowing to rank all approved drugs based on their likely efficacy for COVID-19 patients, aggregate all predictions, and, thereby to arrive at 81 promising repurposing candidates. We validate the accuracy of our predictions using drugs currently in clinical trials, and an expression-based validation of selected candidates suggests that these drugs, with known toxicities and side effects, could be moved to clinical trials rapidly.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge