Xia Dong
Divergent Paths: Separating Homophilic and Heterophilic Learning for Enhanced Graph-level Representations
Apr 06, 2025Abstract:Graph Convolutional Networks (GCNs) are predominantly tailored for graphs displaying homophily, where similar nodes connect, but often fail on heterophilic graphs. The strategy of adopting distinct approaches to learn from homophilic and heterophilic components in node-level tasks has been widely discussed and proven effective both theoretically and experimentally. However, in graph-level tasks, research on this topic remains notably scarce. Addressing this gap, our research conducts an analysis on graphs with nodes' category ID available, distinguishing intra-category and inter-category components as embodiment of homophily and heterophily, respectively. We find while GCNs excel at extracting information within categories, they frequently capture noise from inter-category components. Consequently, it is crucial to employ distinct learning strategies for intra- and inter-category elements. To alleviate this problem, we separately learn the intra- and inter-category parts by a combination of an intra-category convolution (IntraNet) and an inter-category high-pass graph convolution (InterNet). Our IntraNet is supported by sophisticated graph preprocessing steps and a novel category-based graph readout function. For the InterNet, we utilize a high-pass filter to amplify the node disparities, enhancing the recognition of details in the high-frequency components. The proposed approach, DivGNN, combines the IntraNet and InterNet with a gated mechanism and substantially improves classification performance on graph-level tasks, surpassing traditional GNN baselines in effectiveness.
BrainOOD: Out-of-distribution Generalizable Brain Network Analysis
Feb 02, 2025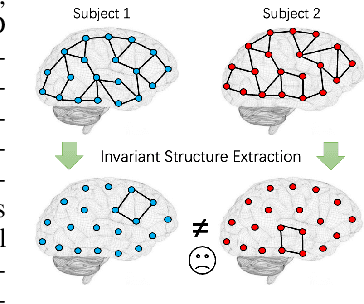
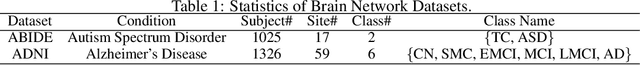
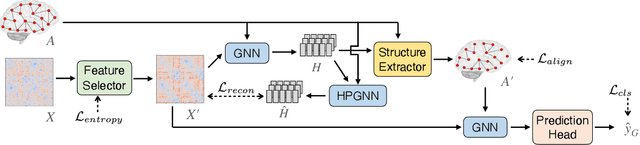
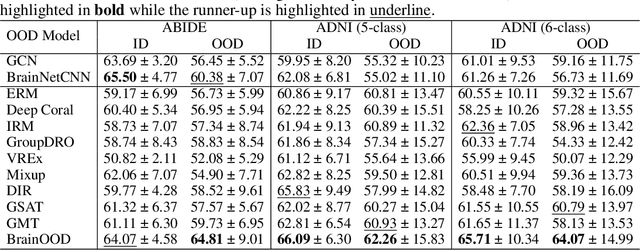
Abstract:In neuroscience, identifying distinct patterns linked to neurological disorders, such as Alzheimer's and Autism, is critical for early diagnosis and effective intervention. Graph Neural Networks (GNNs) have shown promising in analyzing brain networks, but there are two major challenges in using GNNs: (1) distribution shifts in multi-site brain network data, leading to poor Out-of-Distribution (OOD) generalization, and (2) limited interpretability in identifying key brain regions critical to neurological disorders. Existing graph OOD methods, while effective in other domains, struggle with the unique characteristics of brain networks. To bridge these gaps, we introduce BrainOOD, a novel framework tailored for brain networks that enhances GNNs' OOD generalization and interpretability. BrainOOD framework consists of a feature selector and a structure extractor, which incorporates various auxiliary losses including an improved Graph Information Bottleneck (GIB) objective to recover causal subgraphs. By aligning structure selection across brain networks and filtering noisy features, BrainOOD offers reliable interpretations of critical brain regions. Our approach outperforms 16 existing methods and improves generalization to OOD subjects by up to 8.5%. Case studies highlight the scientific validity of the patterns extracted, which aligns with the findings in known neuroscience literature. We also propose the first OOD brain network benchmark, which provides a foundation for future research in this field. Our code is available at https://github.com/AngusMonroe/BrainOOD.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge