Winnie Chiu-Wing Chu
Adapting Vision-Language Foundation Model for Next Generation Medical Ultrasound Image Analysis
Jun 11, 2025Abstract:Medical ultrasonography is an essential imaging technique for examining superficial organs and tissues, including lymph nodes, breast, and thyroid. It employs high-frequency ultrasound waves to generate detailed images of the internal structures of the human body. However, manually contouring regions of interest in these images is a labor-intensive task that demands expertise and often results in inconsistent interpretations among individuals. Vision-language foundation models, which have excelled in various computer vision applications, present new opportunities for enhancing ultrasound image analysis. Yet, their performance is hindered by the significant differences between natural and medical imaging domains. This research seeks to overcome these challenges by developing domain adaptation methods for vision-language foundation models. In this study, we explore the fine-tuning pipeline for vision-language foundation models by utilizing large language model as text refiner with special-designed adaptation strategies and task-driven heads. Our approach has been extensively evaluated on six ultrasound datasets and two tasks: segmentation and classification. The experimental results show that our method can effectively improve the performance of vision-language foundation models for ultrasound image analysis, and outperform the existing state-of-the-art vision-language and pure foundation models. The source code of this study is available at https://github.com/jinggqu/NextGen-UIA.
The Application of Deep Learning for Lymph Node Segmentation: A Systematic Review
May 09, 2025Abstract:Automatic lymph node segmentation is the cornerstone for advances in computer vision tasks for early detection and staging of cancer. Traditional segmentation methods are constrained by manual delineation and variability in operator proficiency, limiting their ability to achieve high accuracy. The introduction of deep learning technologies offers new possibilities for improving the accuracy of lymph node image analysis. This study evaluates the application of deep learning in lymph node segmentation and discusses the methodologies of various deep learning architectures such as convolutional neural networks, encoder-decoder networks, and transformers in analyzing medical imaging data across different modalities. Despite the advancements, it still confronts challenges like the shape diversity of lymph nodes, the scarcity of accurately labeled datasets, and the inadequate development of methods that are robust and generalizable across different imaging modalities. To the best of our knowledge, this is the first study that provides a comprehensive overview of the application of deep learning techniques in lymph node segmentation task. Furthermore, this study also explores potential future research directions, including multimodal fusion techniques, transfer learning, and the use of large-scale pre-trained models to overcome current limitations while enhancing cancer diagnosis and treatment planning strategies.
Chemical Shift Encoding based Double Bonds Quantification in Triglycerides using Deep Image Prior
Jul 02, 2024



Abstract:This study evaluated a deep learning-based method using Deep Image Prior (DIP) to quantify triglyceride double bonds from chemical-shift encoded multi-echo gradient echo images without network training. We employed a cost function based on signal constraints to iteratively update the neural network on a single dataset. The method was validated using phantom experiments and in vivo scans. Results showed close alignment between measured and reference double bond values, with phantom experiments yielding a Pearson correlation coefficient of 0.96 (p = .0005). In vivo results demonstrated good agreement in subcutaneous fat. We conclude that Deep Image Prior shows feasibility for quantifying double bonds and fatty acid content from chemical-shift encoded multi-echo MRI.
Automatic Ultrasound Curve Angle Measurement via Affinity Clustering for Adolescent Idiopathic Scoliosis Evaluation
May 07, 2024



Abstract:The current clinical gold standard for evaluating adolescent idiopathic scoliosis (AIS) is X-ray radiography, using Cobb angle measurement. However, the frequent monitoring of the AIS progression using X-rays poses a challenge due to the cumulative radiation exposure. Although 3D ultrasound has been validated as a reliable and radiation-free alternative for scoliosis assessment, the process of measuring spinal curvature is still carried out manually. Consequently, there is a considerable demand for a fully automatic system that can locate bony landmarks and perform angle measurements. To this end, we introduce an estimation model for automatic ultrasound curve angle (UCA) measurement. The model employs a dual-branch network to detect candidate landmarks and perform vertebra segmentation on ultrasound coronal images. An affinity clustering strategy is utilized within the vertebral segmentation area to illustrate the affinity relationship between candidate landmarks. Subsequently, we can efficiently perform line delineation from a clustered affinity map for UCA measurement. As our method is specifically designed for UCA calculation, this method outperforms other state-of-the-art methods for landmark and line detection tasks. The high correlation between the automatic UCA and Cobb angle (R$^2$=0.858) suggests that our proposed method can potentially replace manual UCA measurement in ultrasound scoliosis assessment.
Uncertainty-Aware Self-supervised Neural Network for Liver $T_{1ρ}$ Mapping with Relaxation Constraint
Jul 07, 2022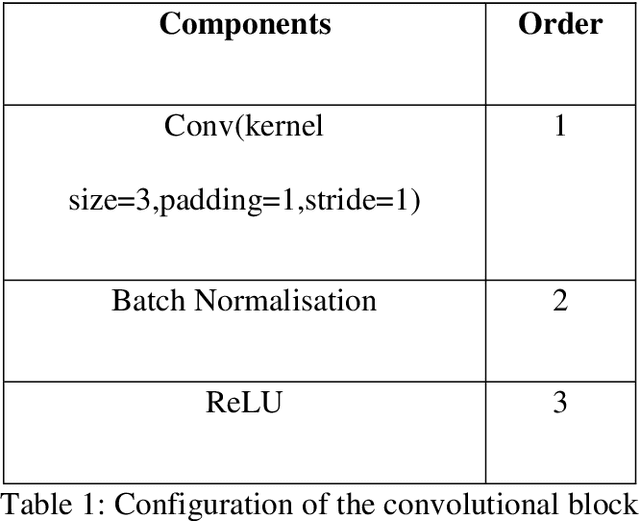

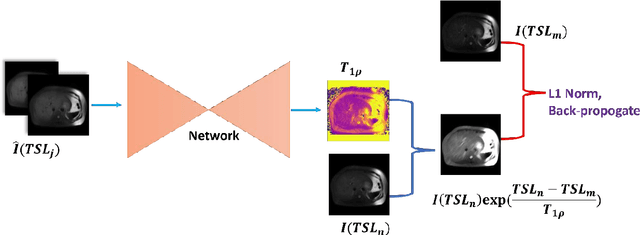
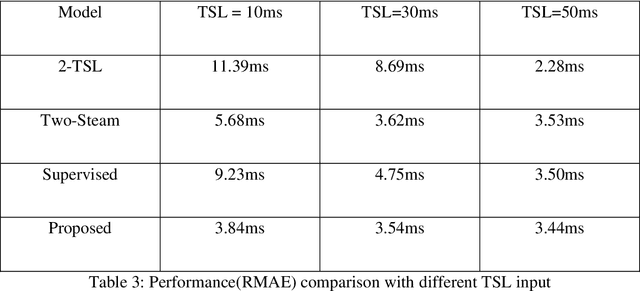
Abstract:$T_{1\rho}$ mapping is a promising quantitative MRI technique for the non-invasive assessment of tissue properties. Learning-based approaches can map $T_{1\rho}$ from a reduced number of $T_{1\rho}$ weighted images, but requires significant amounts of high quality training data. Moreover, existing methods do not provide the confidence level of the $T_{1\rho}$ estimation. To address these problems, we proposed a self-supervised learning neural network that learns a $T_{1\rho}$ mapping using the relaxation constraint in the learning process. Epistemic uncertainty and aleatoric uncertainty are modelled for the $T_{1\rho}$ quantification network to provide a Bayesian confidence estimation of the $T_{1\rho}$ mapping. The uncertainty estimation can also regularize the model to prevent it from learning imperfect data. We conducted experiments on $T_{1\rho}$ data collected from 52 patients with non-alcoholic fatty liver disease. The results showed that our method outperformed the existing methods for $T_{1\rho}$ quantification of the liver using as few as two $T_{1\rho}$-weighted images. Our uncertainty estimation provided a feasible way of modelling the confidence of the self-supervised learning based $T_{1\rho}$ estimation, which is consistent with the reality in liver $T_{1\rho}$ imaging.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge