Wenfang Yao
Addressing Asynchronicity in Clinical Multimodal Fusion via Individualized Chest X-ray Generation
Oct 23, 2024



Abstract:Integrating multi-modal clinical data, such as electronic health records (EHR) and chest X-ray images (CXR), is particularly beneficial for clinical prediction tasks. However, in a temporal setting, multi-modal data are often inherently asynchronous. EHR can be continuously collected but CXR is generally taken with a much longer interval due to its high cost and radiation dose. When clinical prediction is needed, the last available CXR image might have been outdated, leading to suboptimal predictions. To address this challenge, we propose DDL-CXR, a method that dynamically generates an up-to-date latent representation of the individualized CXR images. Our approach leverages latent diffusion models for patient-specific generation strategically conditioned on a previous CXR image and EHR time series, providing information regarding anatomical structures and disease progressions, respectively. In this way, the interaction across modalities could be better captured by the latent CXR generation process, ultimately improving the prediction performance. Experiments using MIMIC datasets show that the proposed model could effectively address asynchronicity in multimodal fusion and consistently outperform existing methods.
DrFuse: Learning Disentangled Representation for Clinical Multi-Modal Fusion with Missing Modality and Modal Inconsistency
Mar 10, 2024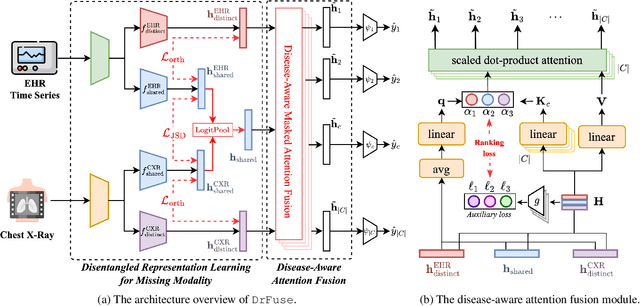
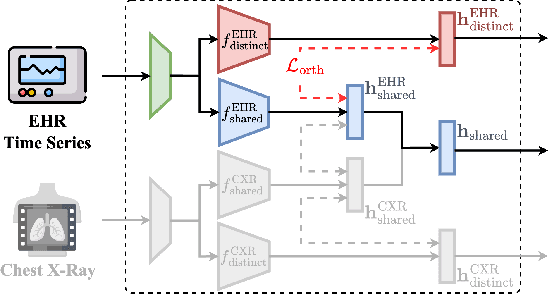
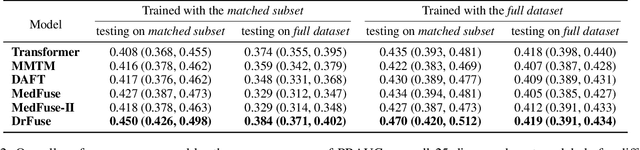
Abstract:The combination of electronic health records (EHR) and medical images is crucial for clinicians in making diagnoses and forecasting prognosis. Strategically fusing these two data modalities has great potential to improve the accuracy of machine learning models in clinical prediction tasks. However, the asynchronous and complementary nature of EHR and medical images presents unique challenges. Missing modalities due to clinical and administrative factors are inevitable in practice, and the significance of each data modality varies depending on the patient and the prediction target, resulting in inconsistent predictions and suboptimal model performance. To address these challenges, we propose DrFuse to achieve effective clinical multi-modal fusion. It tackles the missing modality issue by disentangling the features shared across modalities and those unique within each modality. Furthermore, we address the modal inconsistency issue via a disease-wise attention layer that produces the patient- and disease-wise weighting for each modality to make the final prediction. We validate the proposed method using real-world large-scale datasets, MIMIC-IV and MIMIC-CXR. Experimental results show that the proposed method significantly outperforms the state-of-the-art models. Our implementation is publicly available at https://github.com/dorothy-yao/drfuse.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge