Vlad Pekar
Virtual Histological Staining of Label-Free Total Absorption Photoacoustic Remote Sensing (TA-PARS)
Apr 01, 2022
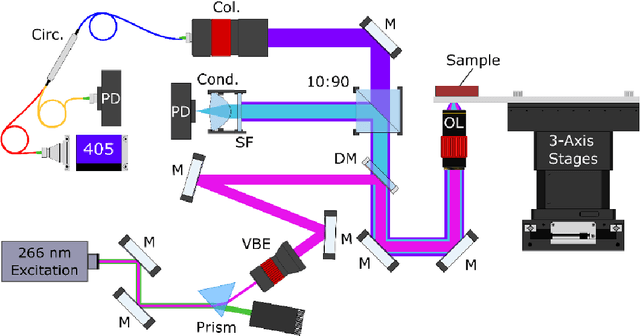
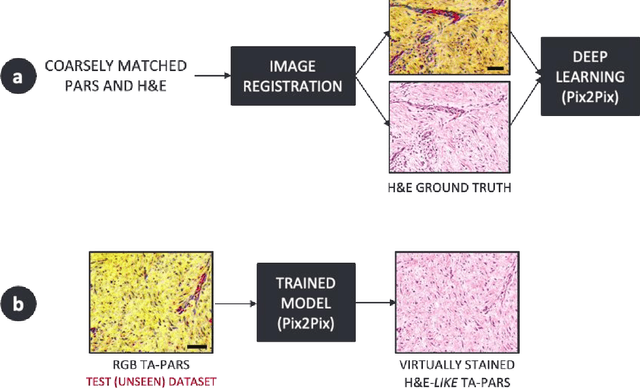

Abstract:Histopathological visualizations are a pillar of modern medicine and biological research. Surgical oncology relies exclusively on post-operative histology to determine definitive surgical success and guide adjuvant treatments. The current histology workflow is based on bright-field microscopic assessment of histochemical stained tissues and has some major limitations. For example, the preparation of stained specimens for brightfield assessment requires lengthy sample processing, delaying interventions for days or even weeks. Hence, there is a pressing need for improved histopathology methods. In this paper, we present a deep-learning-based approach for virtual label-free histochemical staining of total-absorption photoacoustic remote sensing (TA-PARS) images of unstained tissue. TA-PARS provides an array of directly measured label-free contrasts such as scattering and total absorption (radiative and non-radiative), ideal for developing H&E colorizations without the need to infer arbitrary tissue structures. We use a Pix2Pix generative adversarial network (GAN) to develop visualizations analogous to H&E staining from label-free TA-PARS images. Thin sections of human skin tissue were first virtually stained with the TA-PARS, then were chemically stained with H&E producing a one-to-one comparison between the virtual and chemical staining. The one-to-one matched virtually- and chemically- stained images exhibit high concordance validating the digital colorization of the TA-PARS images against the gold standard H&E. TA-PARS images were reviewed by four dermatologic pathologists who confirmed they are of diagnostic quality, and that resolution, contrast, and color permitted interpretation as if they were H&E. The presented approach paves the way for the development of TA-PARS slide-free histology, which promises to dramatically reduce the time from specimen resection to histological imaging.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge