Vivek Vajipey
Tony
Humanity's Last Exam
Jan 24, 2025Abstract:Benchmarks are important tools for tracking the rapid advancements in large language model (LLM) capabilities. However, benchmarks are not keeping pace in difficulty: LLMs now achieve over 90\% accuracy on popular benchmarks like MMLU, limiting informed measurement of state-of-the-art LLM capabilities. In response, we introduce Humanity's Last Exam (HLE), a multi-modal benchmark at the frontier of human knowledge, designed to be the final closed-ended academic benchmark of its kind with broad subject coverage. HLE consists of 3,000 questions across dozens of subjects, including mathematics, humanities, and the natural sciences. HLE is developed globally by subject-matter experts and consists of multiple-choice and short-answer questions suitable for automated grading. Each question has a known solution that is unambiguous and easily verifiable, but cannot be quickly answered via internet retrieval. State-of-the-art LLMs demonstrate low accuracy and calibration on HLE, highlighting a significant gap between current LLM capabilities and the expert human frontier on closed-ended academic questions. To inform research and policymaking upon a clear understanding of model capabilities, we publicly release HLE at https://lastexam.ai.
CriticAL: Critic Automation with Language Models
Nov 10, 2024Abstract:Understanding the world through models is a fundamental goal of scientific research. While large language model (LLM) based approaches show promise in automating scientific discovery, they often overlook the importance of criticizing scientific models. Criticizing models deepens scientific understanding and drives the development of more accurate models. Automating model criticism is difficult because it traditionally requires a human expert to define how to compare a model with data and evaluate if the discrepancies are significant--both rely heavily on understanding the modeling assumptions and domain. Although LLM-based critic approaches are appealing, they introduce new challenges: LLMs might hallucinate the critiques themselves. Motivated by this, we introduce CriticAL (Critic Automation with Language Models). CriticAL uses LLMs to generate summary statistics that capture discrepancies between model predictions and data, and applies hypothesis tests to evaluate their significance. We can view CriticAL as a verifier that validates models and their critiques by embedding them in a hypothesis testing framework. In experiments, we evaluate CriticAL across key quantitative and qualitative dimensions. In settings where we synthesize discrepancies between models and datasets, CriticAL reliably generates correct critiques without hallucinating incorrect ones. We show that both human and LLM judges consistently prefer CriticAL's critiques over alternative approaches in terms of transparency and actionability. Finally, we show that CriticAL's critiques enable an LLM scientist to improve upon human-designed models on real-world datasets.
Annual field-scale maps of tall and short crops at the global scale using GEDI and Sentinel-2
Dec 19, 2022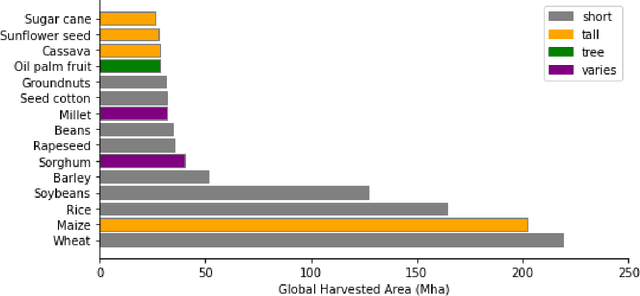
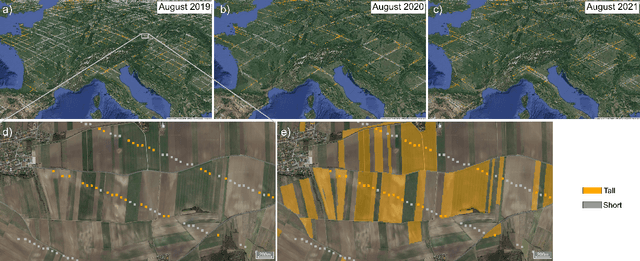
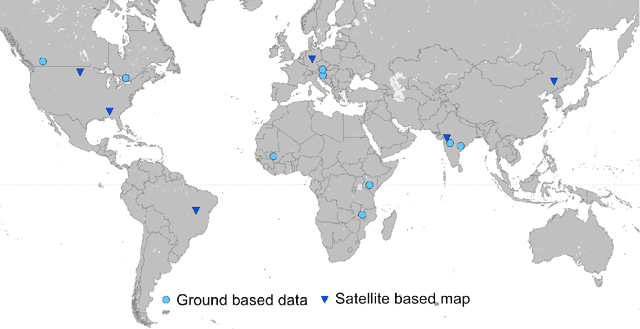
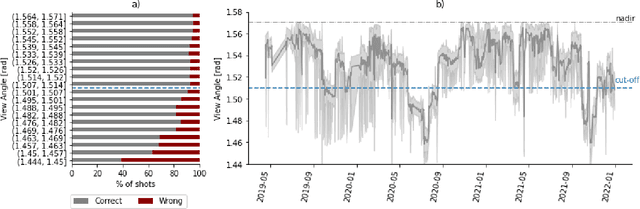
Abstract:Crop type maps are critical for tracking agricultural land use and estimating crop production. Remote sensing has proven an efficient and reliable tool for creating these maps in regions with abundant ground labels for model training, yet these labels remain difficult to obtain in many regions and years. NASA's Global Ecosystem Dynamics Investigation (GEDI) spaceborne lidar instrument, originally designed for forest monitoring, has shown promise for distinguishing tall and short crops. In the current study, we leverage GEDI to develop wall-to-wall maps of short vs tall crops on a global scale at 10 m resolution for 2019-2021. Specifically, we show that (1) GEDI returns can reliably be classified into tall and short crops after removing shots with extreme view angles or topographic slope, (2) the frequency of tall crops over time can be used to identify months when tall crops are at their peak height, and (3) GEDI shots in these months can then be used to train random forest models that use Sentinel-2 time series to accurately predict short vs. tall crops. Independent reference data from around the world are then used to evaluate these GEDI-S2 maps. We find that GEDI-S2 performed nearly as well as models trained on thousands of local reference training points, with accuracies of at least 87% and often above 90% throughout the Americas, Europe, and East Asia. Systematic underestimation of tall crop area was observed in regions where crops frequently exhibit low biomass, namely Africa and South Asia, and further work is needed in these systems. Although the GEDI-S2 approach only differentiates tall from short crops, in many landscapes this distinction goes a long way toward mapping the main individual crop types. The combination of GEDI and Sentinel-2 thus presents a very promising path towards global crop mapping with minimal reliance on ground data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge