Vijaya B. Kolachalama
Higher-Order Domain Generalization in Magnetic Resonance-Based Assessment of Alzheimer's Disease
Jan 04, 2026Abstract:Despite progress in deep learning for Alzheimer's disease (AD) diagnostics, models trained on structural magnetic resonance imaging (sMRI) often do not perform well when applied to new cohorts due to domain shifts from varying scanners, protocols and patient demographics. AD, the primary driver of dementia, manifests through progressive cognitive and neuroanatomical changes like atrophy and ventricular expansion, making robust, generalizable classification essential for real-world use. While convolutional neural networks and transformers have advanced feature extraction via attention and fusion techniques, single-domain generalization (SDG) remains underexplored yet critical, given the fragmented nature of AD datasets. To bridge this gap, we introduce Extended MixStyle (EM), a framework for blending higher-order feature moments (skewness and kurtosis) to mimic diverse distributional variations. Trained on sMRI data from the National Alzheimer's Coordinating Center (NACC; n=4,647) to differentiate persons with normal cognition (NC) from those with mild cognitive impairment (MCI) or AD and tested on three unseen cohorts (total n=3,126), EM yields enhanced cross-domain performance, improving macro-F1 on average by 2.4 percentage points over state-of-the-art SDG benchmarks, underscoring its promise for invariant, reliable AD detection in heterogeneous real-world settings. The source code will be made available upon acceptance at https://github.com/zobia111/Extended-Mixstyle.
A graph-transformer for whole slide image classification
May 19, 2022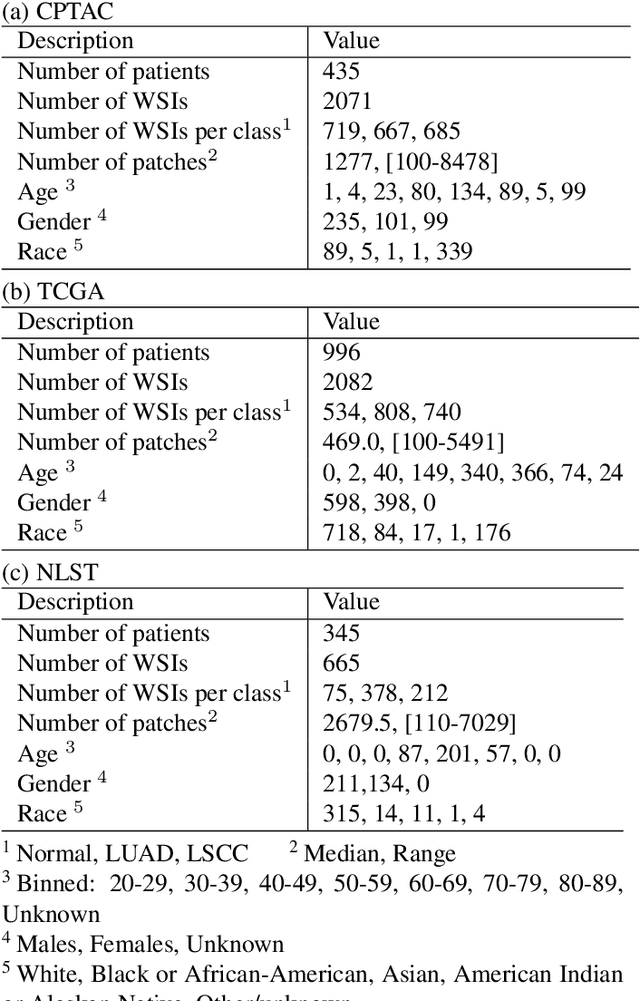
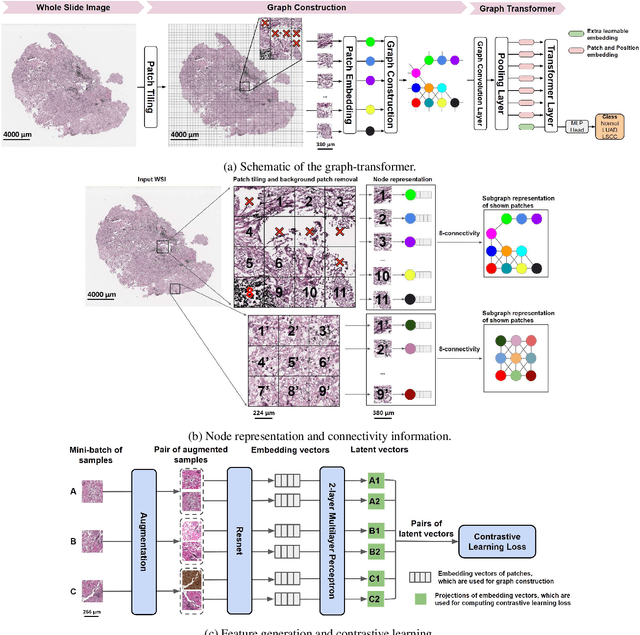
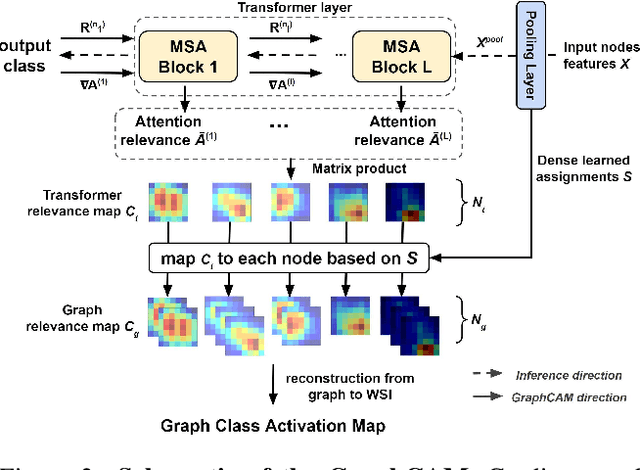
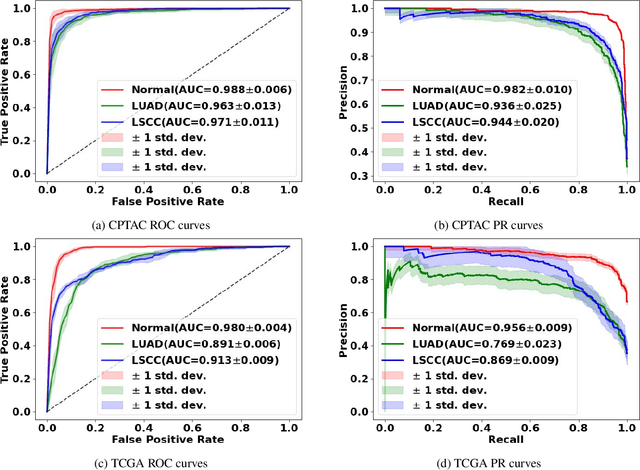
Abstract:Deep learning is a powerful tool for whole slide image (WSI) analysis. Typically, when performing supervised deep learning, a WSI is divided into small patches, trained and the outcomes are aggregated to estimate disease grade. However, patch-based methods introduce label noise during training by assuming that each patch is independent with the same label as the WSI and neglect overall WSI-level information that is significant in disease grading. Here we present a Graph-Transformer (GT) that fuses a graph-based representation of an WSI and a vision transformer for processing pathology images, called GTP, to predict disease grade. We selected $4,818$ WSIs from the Clinical Proteomic Tumor Analysis Consortium (CPTAC), the National Lung Screening Trial (NLST), and The Cancer Genome Atlas (TCGA), and used GTP to distinguish adenocarcinoma (LUAD) and squamous cell carcinoma (LSCC) from adjacent non-cancerous tissue (normal). First, using NLST data, we developed a contrastive learning framework to generate a feature extractor. This allowed us to compute feature vectors of individual WSI patches, which were used to represent the nodes of the graph followed by construction of the GTP framework. Our model trained on the CPTAC data achieved consistently high performance on three-label classification (normal versus LUAD versus LSCC: mean accuracy$= 91.2$ $\pm$ $2.5\%$) based on five-fold cross-validation, and mean accuracy $= 82.3$ $\pm$ $1.0\%$ on external test data (TCGA). We also introduced a graph-based saliency mapping technique, called GraphCAM, that can identify regions that are highly associated with the class label. Our findings demonstrate GTP as an interpretable and effective deep learning framework for WSI-level classification.
U-Net-and-a-half: Convolutional network for biomedical image segmentation using multiple expert-driven annotations
Aug 10, 2021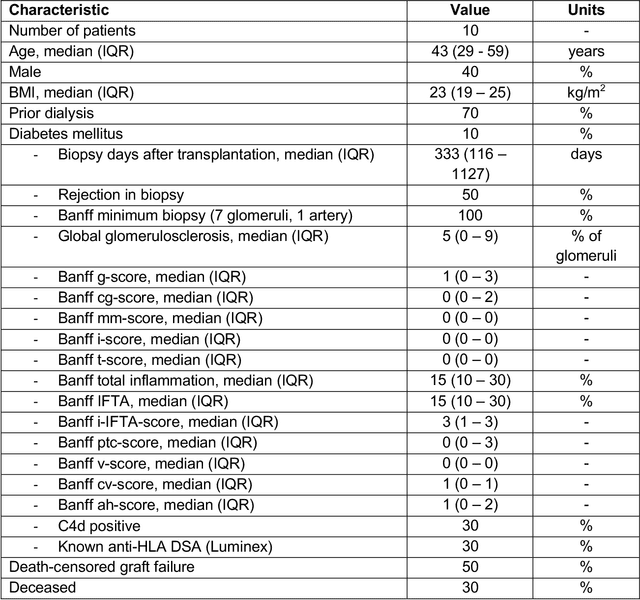
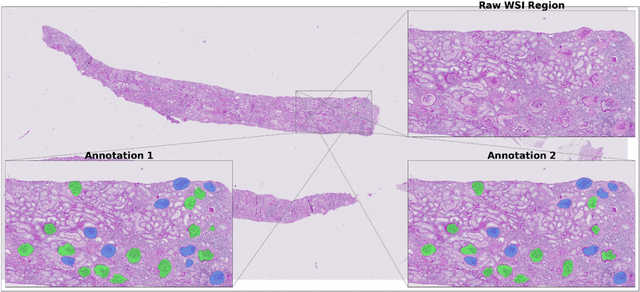
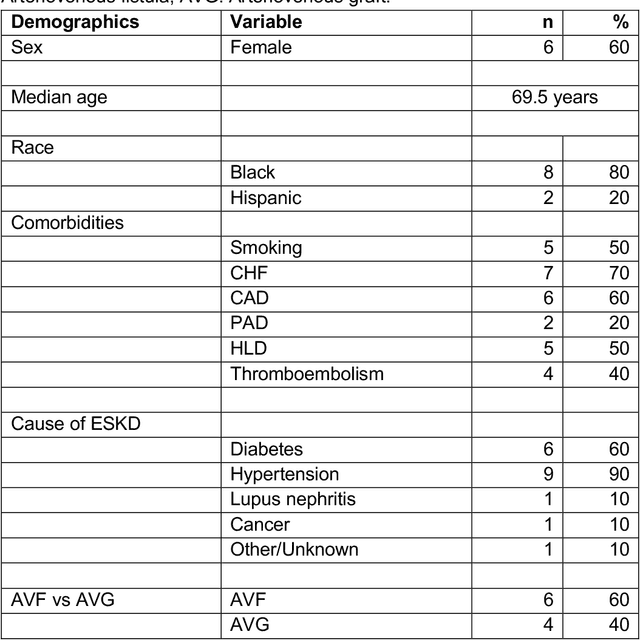
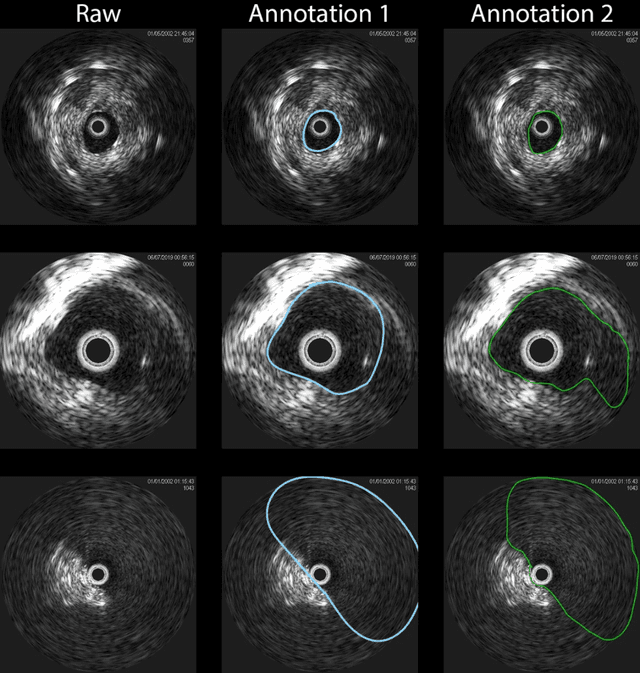
Abstract:Development of deep learning systems for biomedical segmentation often requires access to expert-driven, manually annotated datasets. If more than a single expert is involved in the annotation of the same images, then the inter-expert agreement is not necessarily perfect, and no single expert annotation can precisely capture the so-called ground truth of the regions of interest on all images. Also, it is not trivial to generate a reference estimate using annotations from multiple experts. Here we present a deep neural network, defined as U-Net-and-a-half, which can simultaneously learn from annotations performed by multiple experts on the same set of images. U-Net-and-a-half contains a convolutional encoder to generate features from the input images, multiple decoders that allow simultaneous learning from image masks obtained from annotations that were independently generated by multiple experts, and a shared low-dimensional feature space. To demonstrate the applicability of our framework, we used two distinct datasets from digital pathology and radiology, respectively. Specifically, we trained two separate models using pathologist-driven annotations of glomeruli on whole slide images of human kidney biopsies (10 patients), and radiologist-driven annotations of lumen cross-sections of human arteriovenous fistulae obtained from intravascular ultrasound images (10 patients), respectively. The models based on U-Net-and-a-half exceeded the performance of the traditional U-Net models trained on single expert annotations alone, thus expanding the scope of multitask learning in the context of biomedical image segmentation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge