Jesper Kers
PathBench-MIL: A Comprehensive AutoML and Benchmarking Framework for Multiple Instance Learning in Histopathology
Dec 19, 2025Abstract:We introduce PathBench-MIL, an open-source AutoML and benchmarking framework for multiple instance learning (MIL) in histopathology. The system automates end-to-end MIL pipeline construction, including preprocessing, feature extraction, and MIL-aggregation, and provides reproducible benchmarking of dozens of MIL models and feature extractors. PathBench-MIL integrates visualization tooling, a unified configuration system, and modular extensibility, enabling rapid experimentation and standardization across datasets and tasks. PathBench-MIL is publicly available at https://github.com/Sbrussee/PathBench-MIL
Graph Neural Networks in Histopathology: Emerging Trends and Future Directions
Jun 18, 2024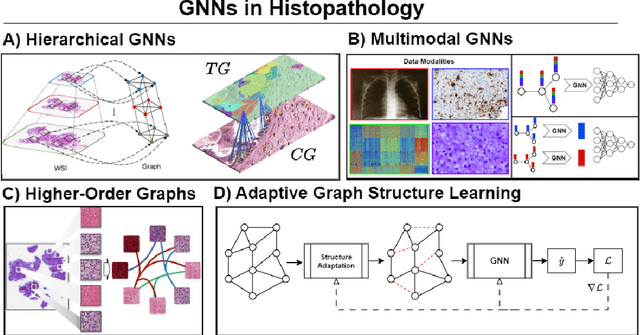
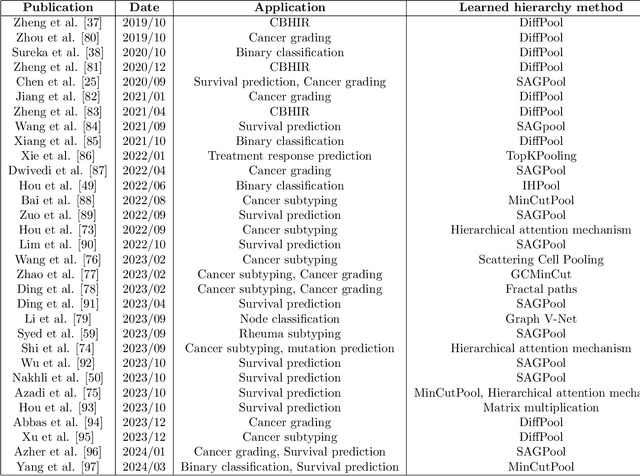
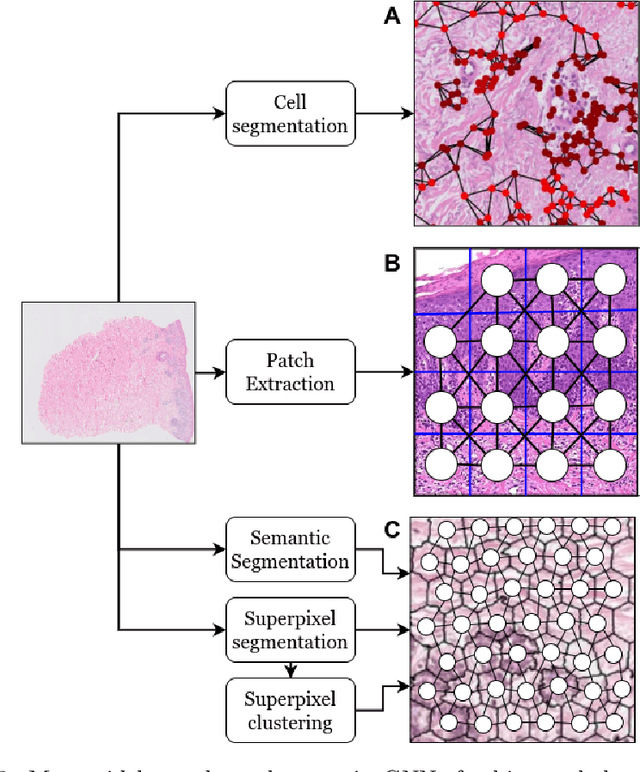
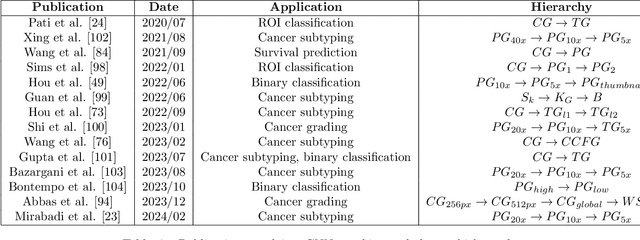
Abstract:Histopathological analysis of Whole Slide Images (WSIs) has seen a surge in the utilization of deep learning methods, particularly Convolutional Neural Networks (CNNs). However, CNNs often fall short in capturing the intricate spatial dependencies inherent in WSIs. Graph Neural Networks (GNNs) present a promising alternative, adept at directly modeling pairwise interactions and effectively discerning the topological tissue and cellular structures within WSIs. Recognizing the pressing need for deep learning techniques that harness the topological structure of WSIs, the application of GNNs in histopathology has experienced rapid growth. In this comprehensive review, we survey GNNs in histopathology, discuss their applications, and exploring emerging trends that pave the way for future advancements in the field. We begin by elucidating the fundamentals of GNNs and their potential applications in histopathology. Leveraging quantitative literature analysis, we identify four emerging trends: Hierarchical GNNs, Adaptive Graph Structure Learning, Multimodal GNNs, and Higher-order GNNs. Through an in-depth exploration of these trends, we offer insights into the evolving landscape of GNNs in histopathological analysis. Based on our findings, we propose future directions to propel the field forward. Our analysis serves to guide researchers and practitioners towards innovative approaches and methodologies, fostering advancements in histopathological analysis through the lens of graph neural networks.
Segmentation of diagnostic tissue compartments on whole slide images with renal thrombotic microangiopathies (TMAs)
Nov 28, 2023



Abstract:The thrombotic microangiopathies (TMAs) manifest in renal biopsy histology with a broad spectrum of acute and chronic findings. Precise diagnostic criteria for a renal biopsy diagnosis of TMA are missing. As a first step towards a machine learning- and computer vision-based analysis of wholes slide images from renal biopsies, we trained a segmentation model for the decisive diagnostic kidney tissue compartments artery, arteriole, glomerulus on a set of whole slide images from renal biopsies with TMAs and Mimickers (distinct diseases with a similar nephropathological appearance as TMA like severe benign nephrosclerosis, various vasculitides, Bevacizumab-plug glomerulopathy, arteriolar light chain deposition disease). Our segmentation model combines a U-Net-based tissue detection with a Shifted windows-transformer architecture to reach excellent segmentation results for even the most severely altered glomeruli, arterioles and arteries, even on unseen staining domains from a different nephropathology lab. With accurate automatic segmentation of the decisive renal biopsy compartments in human renal vasculopathies, we have laid the foundation for large-scale compartment-specific machine learning and computer vision analysis of renal biopsy repositories with TMAs.
Advances in Kidney Biopsy Structural Assessment through Dense Instance Segmentation
Sep 29, 2023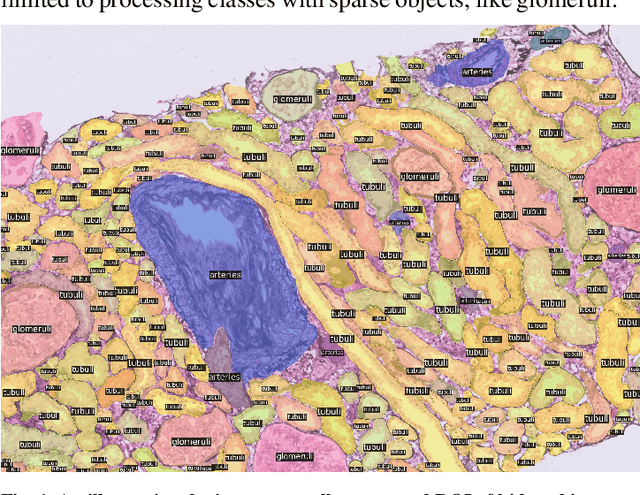
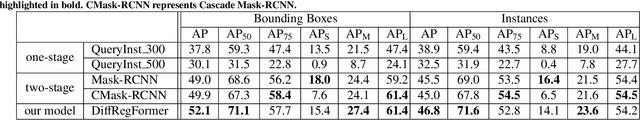
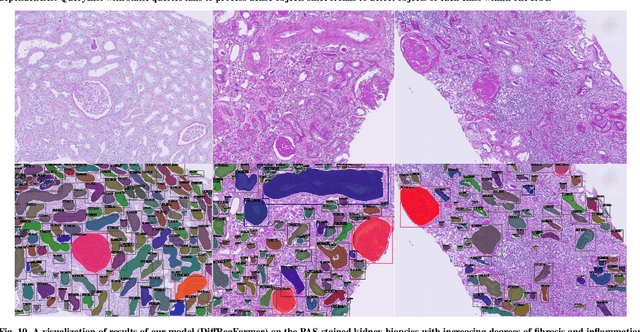
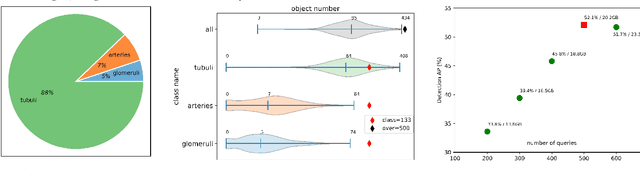
Abstract:The kidney biopsy is the gold standard for the diagnosis of kidney diseases. Lesion scores made by expert renal pathologists are semi-quantitative and suffer from high inter-observer variability. Automatically obtaining statistics per segmented anatomical object, therefore, can bring significant benefits in reducing labor and this inter-observer variability. Instance segmentation for a biopsy, however, has been a challenging problem due to (a) the on average large number (around 300 to 1000) of densely touching anatomical structures, (b) with multiple classes (at least 3) and (c) in different sizes and shapes. The currently used instance segmentation models cannot simultaneously deal with these challenges in an efficient yet generic manner. In this paper, we propose the first anchor-free instance segmentation model that combines diffusion models, transformer modules, and RCNNs (regional convolution neural networks). Our model is trained on just one NVIDIA GeForce RTX 3090 GPU, but can efficiently recognize more than 500 objects with 3 common anatomical object classes in renal biopsies, i.e., glomeruli, tubuli, and arteries. Our data set consisted of 303 patches extracted from 148 Jones' silver-stained renal whole slide images (WSIs), where 249 patches were used for training and 54 patches for evaluation. In addition, without adjustment or retraining, the model can directly transfer its domain to generate decent instance segmentation results from PAS-stained WSIs. Importantly, it outperforms other baseline models and reaches an AP 51.7% in detection as the new state-of-the-art.
U-Net-and-a-half: Convolutional network for biomedical image segmentation using multiple expert-driven annotations
Aug 10, 2021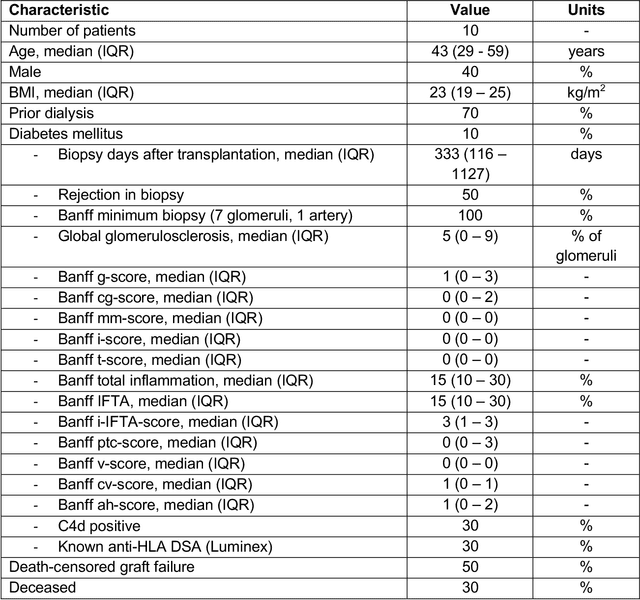
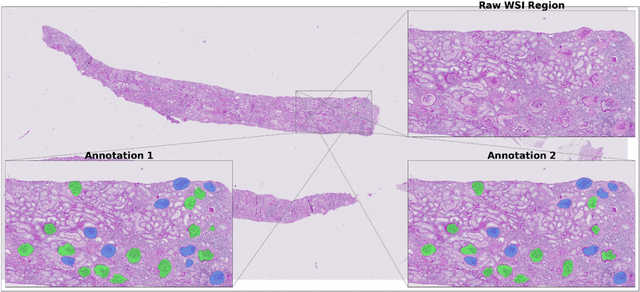
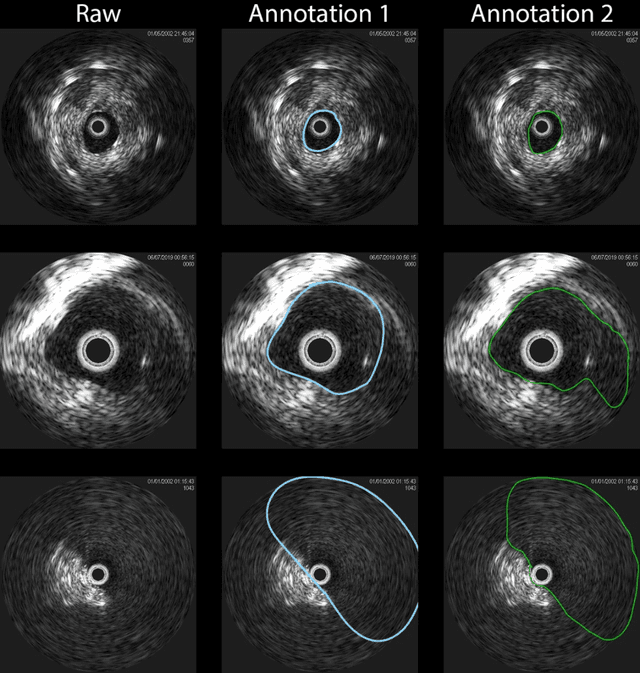
Abstract:Development of deep learning systems for biomedical segmentation often requires access to expert-driven, manually annotated datasets. If more than a single expert is involved in the annotation of the same images, then the inter-expert agreement is not necessarily perfect, and no single expert annotation can precisely capture the so-called ground truth of the regions of interest on all images. Also, it is not trivial to generate a reference estimate using annotations from multiple experts. Here we present a deep neural network, defined as U-Net-and-a-half, which can simultaneously learn from annotations performed by multiple experts on the same set of images. U-Net-and-a-half contains a convolutional encoder to generate features from the input images, multiple decoders that allow simultaneous learning from image masks obtained from annotations that were independently generated by multiple experts, and a shared low-dimensional feature space. To demonstrate the applicability of our framework, we used two distinct datasets from digital pathology and radiology, respectively. Specifically, we trained two separate models using pathologist-driven annotations of glomeruli on whole slide images of human kidney biopsies (10 patients), and radiologist-driven annotations of lumen cross-sections of human arteriovenous fistulae obtained from intravascular ultrasound images (10 patients), respectively. The models based on U-Net-and-a-half exceeded the performance of the traditional U-Net models trained on single expert annotations alone, thus expanding the scope of multitask learning in the context of biomedical image segmentation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge