Tri Q. Nguyen
Pathologist-like explainable AI for interpretable Gleason grading in prostate cancer
Oct 19, 2024



Abstract:The aggressiveness of prostate cancer, the most common cancer in men worldwide, is primarily assessed based on histopathological data using the Gleason scoring system. While artificial intelligence (AI) has shown promise in accurately predicting Gleason scores, these predictions often lack inherent explainability, potentially leading to distrust in human-machine interactions. To address this issue, we introduce a novel dataset of 1,015 tissue microarray core images, annotated by an international group of 54 pathologists. The annotations provide detailed localized pattern descriptions for Gleason grading in line with international guidelines. Utilizing this dataset, we develop an inherently explainable AI system based on a U-Net architecture that provides predictions leveraging pathologists' terminology. This approach circumvents post-hoc explainability methods while maintaining or exceeding the performance of methods trained directly for Gleason pattern segmentation (Dice score: 0.713 $\pm$ 0.003 trained on explanations vs. 0.691 $\pm$ 0.010 trained on Gleason patterns). By employing soft labels during training, we capture the intrinsic uncertainty in the data, yielding strong results in Gleason pattern segmentation even in the context of high interobserver variability. With the release of this dataset, we aim to encourage further research into segmentation in medical tasks with high levels of subjectivity and to advance the understanding of pathologists' reasoning processes.
Advances in Kidney Biopsy Structural Assessment through Dense Instance Segmentation
Sep 29, 2023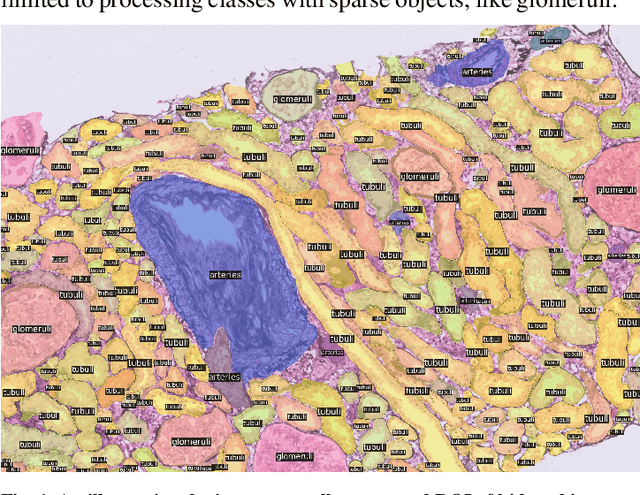
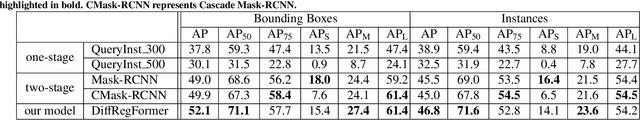
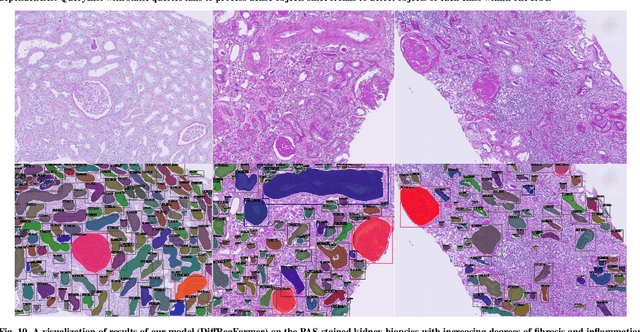
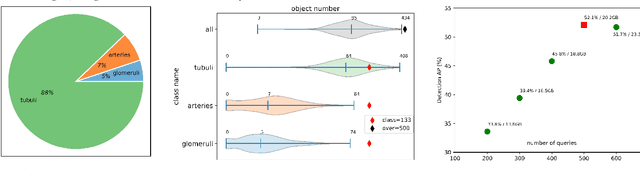
Abstract:The kidney biopsy is the gold standard for the diagnosis of kidney diseases. Lesion scores made by expert renal pathologists are semi-quantitative and suffer from high inter-observer variability. Automatically obtaining statistics per segmented anatomical object, therefore, can bring significant benefits in reducing labor and this inter-observer variability. Instance segmentation for a biopsy, however, has been a challenging problem due to (a) the on average large number (around 300 to 1000) of densely touching anatomical structures, (b) with multiple classes (at least 3) and (c) in different sizes and shapes. The currently used instance segmentation models cannot simultaneously deal with these challenges in an efficient yet generic manner. In this paper, we propose the first anchor-free instance segmentation model that combines diffusion models, transformer modules, and RCNNs (regional convolution neural networks). Our model is trained on just one NVIDIA GeForce RTX 3090 GPU, but can efficiently recognize more than 500 objects with 3 common anatomical object classes in renal biopsies, i.e., glomeruli, tubuli, and arteries. Our data set consisted of 303 patches extracted from 148 Jones' silver-stained renal whole slide images (WSIs), where 249 patches were used for training and 54 patches for evaluation. In addition, without adjustment or retraining, the model can directly transfer its domain to generate decent instance segmentation results from PAS-stained WSIs. Importantly, it outperforms other baseline models and reaches an AP 51.7% in detection as the new state-of-the-art.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge