Vidhiwar Singh Rathour
Deep Reinforcement Learning in Computer Vision: A Comprehensive Survey
Aug 25, 2021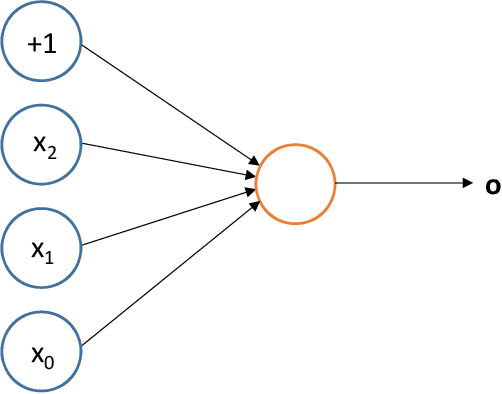
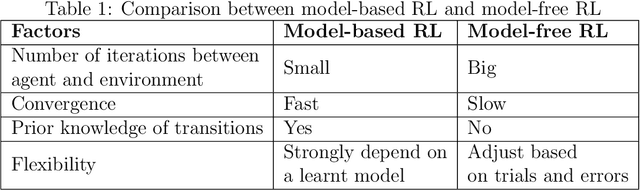
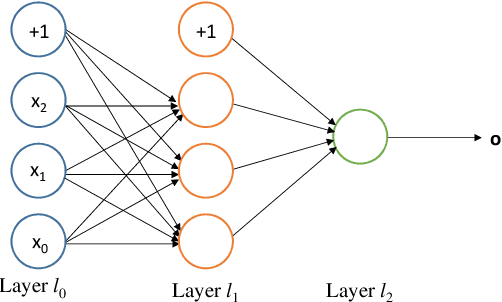
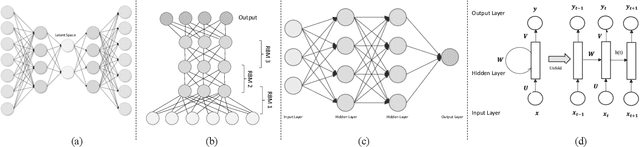
Abstract:Deep reinforcement learning augments the reinforcement learning framework and utilizes the powerful representation of deep neural networks. Recent works have demonstrated the remarkable successes of deep reinforcement learning in various domains including finance, medicine, healthcare, video games, robotics, and computer vision. In this work, we provide a detailed review of recent and state-of-the-art research advances of deep reinforcement learning in computer vision. We start with comprehending the theories of deep learning, reinforcement learning, and deep reinforcement learning. We then propose a categorization of deep reinforcement learning methodologies and discuss their advantages and limitations. In particular, we divide deep reinforcement learning into seven main categories according to their applications in computer vision, i.e. (i)landmark localization (ii) object detection; (iii) object tracking; (iv) registration on both 2D image and 3D image volumetric data (v) image segmentation; (vi) videos analysis; and (vii) other applications. Each of these categories is further analyzed with reinforcement learning techniques, network design, and performance. Moreover, we provide a comprehensive analysis of the existing publicly available datasets and examine source code availability. Finally, we present some open issues and discuss future research directions on deep reinforcement learning in computer vision
Roughness Index and Roughness Distance for Benchmarking Medical Segmentation
Mar 23, 2021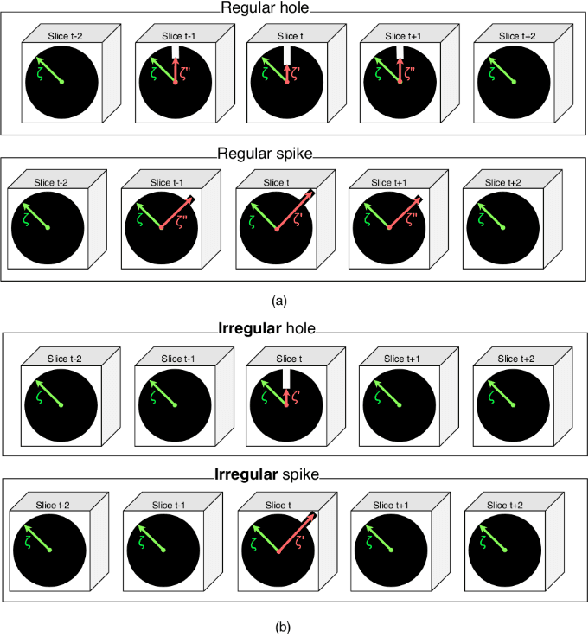
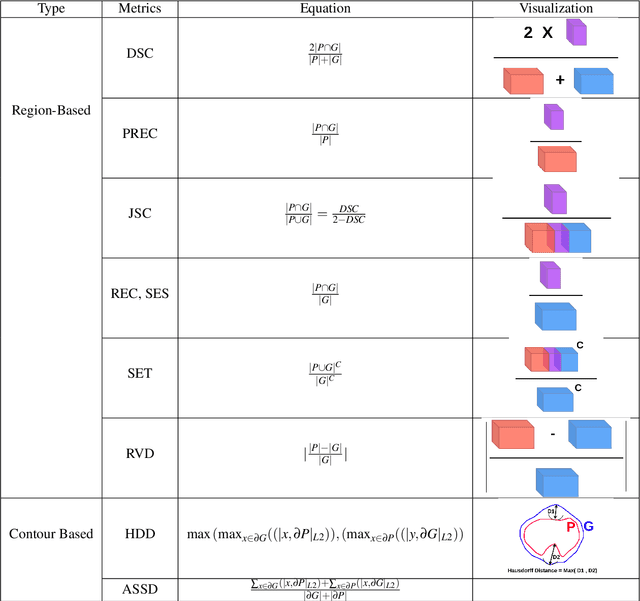
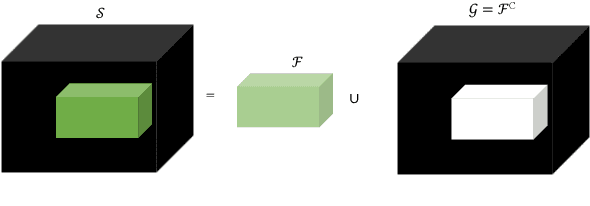

Abstract:Medical image segmentation is one of the most challenging tasks in medical image analysis and has been widely developed for many clinical applications. Most of the existing metrics have been first designed for natural images and then extended to medical images. While object surface plays an important role in medical segmentation and quantitative analysis i.e. analyze brain tumor surface, measure gray matter volume, most of the existing metrics are limited when it comes to analyzing the object surface, especially to tell about surface smoothness or roughness of a given volumetric object or to analyze the topological errors. In this paper, we first analysis both pros and cons of all existing medical image segmentation metrics, specially on volumetric data. We then propose an appropriate roughness index and roughness distance for medical image segmentation analysis and evaluation. Our proposed method addresses two kinds of segmentation errors, i.e. (i)topological errors on boundary/surface and (ii)irregularities on the boundary/surface. The contribution of this work is four-fold: (i) detect irregular spikes/holes on a surface, (ii) propose roughness index to measure surface roughness of a given object, (iii) propose a roughness distance to measure the distance of two boundaries/surfaces by utilizing the proposed roughness index and (iv) suggest an algorithm which helps to remove the irregular spikes/holes to smooth the surface. Our proposed roughness index and roughness distance are built upon the solid surface roughness parameter which has been successfully developed in the civil engineering.
Invertible Residual Network with Regularization for Effective Medical Image Segmentation
Mar 16, 2021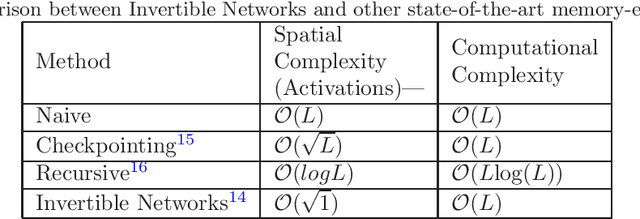
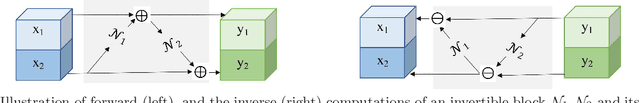
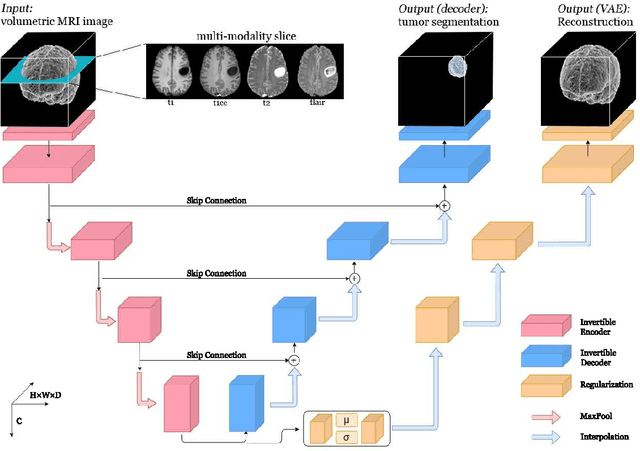
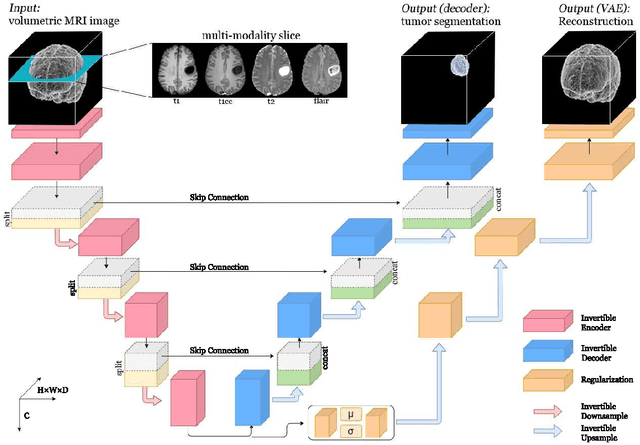
Abstract:Deep Convolutional Neural Networks (CNNs) i.e. Residual Networks (ResNets) have been used successfully for many computer vision tasks, but are difficult to scale to 3D volumetric medical data. Memory is increasingly often the bottleneck when training 3D Convolutional Neural Networks (CNNs). Recently, invertible neural networks have been applied to significantly reduce activation memory footprint when training neural networks with backpropagation thanks to the invertible functions that allow retrieving its input from its output without storing intermediate activations in memory to perform the backpropagation. Among many successful network architectures, 3D Unet has been established as a standard architecture for volumetric medical segmentation. Thus, we choose 3D Unet as a baseline for a non-invertible network and we then extend it with the invertible residual network. In this paper, we proposed two versions of the invertible Residual Network, namely Partially Invertible Residual Network (Partially-InvRes) and Fully Invertible Residual Network (Fully-InvRes). In Partially-InvRes, the invertible residual layer is defined by a technique called additive coupling whereas in Fully-InvRes, both invertible upsampling and downsampling operations are learned based on squeezing (known as pixel shuffle). Furthermore, to avoid the overfitting problem because of less training data, a variational auto-encoder (VAE) branch is added to reconstruct the input volumetric data itself. Our results indicate that by using partially/fully invertible networks as the central workhorse in volumetric segmentation, we not only reduce memory overhead but also achieve compatible segmentation performance compared against the non-invertible 3D Unet. We have demonstrated the proposed networks on various volumetric datasets such as iSeg 2019 and BraTS 2020.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge