Vedant Sanil
AQuA: A Benchmarking Tool for Label Quality Assessment
Jun 15, 2023
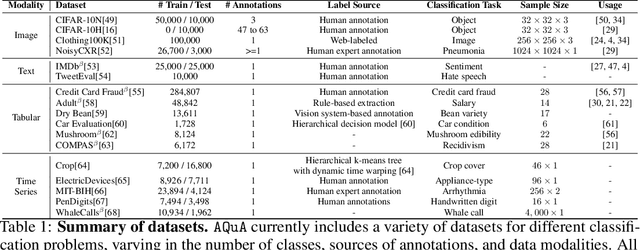
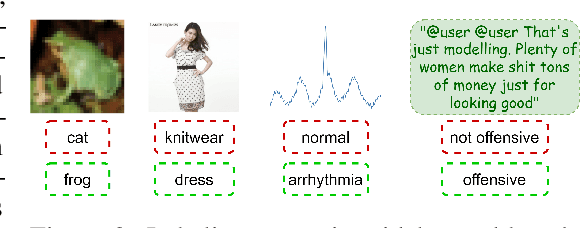
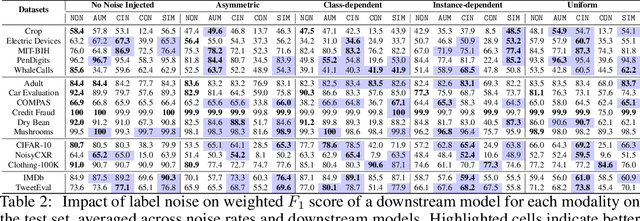
Abstract:Machine learning (ML) models are only as good as the data they are trained on. But recent studies have found datasets widely used to train and evaluate ML models, e.g. ImageNet, to have pervasive labeling errors. Erroneous labels on the train set hurt ML models' ability to generalize, and they impact evaluation and model selection using the test set. Consequently, learning in the presence of labeling errors is an active area of research, yet this field lacks a comprehensive benchmark to evaluate these methods. Most of these methods are evaluated on a few computer vision datasets with significant variance in the experimental protocols. With such a large pool of methods and inconsistent evaluation, it is also unclear how ML practitioners can choose the right models to assess label quality in their data. To this end, we propose a benchmarking environment AQuA to rigorously evaluate methods that enable machine learning in the presence of label noise. We also introduce a design space to delineate concrete design choices of label error detection models. We hope that our proposed design space and benchmark enable practitioners to choose the right tools to improve their label quality and that our benchmark enables objective and rigorous evaluation of machine learning tools facing mislabeled data.
Recovering Sparse and Interpretable Subgroups with Heterogeneous Treatment Effects with Censored Time-to-Event Outcomes
Feb 24, 2023Abstract:Studies involving both randomized experiments as well as observational data typically involve time-to-event outcomes such as time-to-failure, death or onset of an adverse condition. Such outcomes are typically subject to censoring due to loss of follow-up and established statistical practice involves comparing treatment efficacy in terms of hazard ratios between the treated and control groups. In this paper we propose a statistical approach to recovering sparse phenogroups (or subtypes) that demonstrate differential treatment effects as compared to the study population. Our approach involves modelling the data as a mixture while enforcing parameter shrinkage through structured sparsity regularization. We propose a novel inference procedure for the proposed model and demonstrate its efficacy in recovering sparse phenotypes across large landmark real world clinical studies in cardiovascular health.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge