Tongyue Shi
Stochastic Parrots or ICU Experts? Large Language Models in Critical Care Medicine: A Scoping Review
Jul 27, 2024Abstract:With the rapid development of artificial intelligence (AI), large language models (LLMs) have shown strong capabilities in natural language understanding, reasoning, and generation, attracting amounts of research interest in applying LLMs to health and medicine. Critical care medicine (CCM) provides diagnosis and treatment for critically ill patients who often require intensive monitoring and interventions in intensive care units (ICUs). Can LLMs be applied to CCM? Are LLMs just like stochastic parrots or ICU experts in assisting clinical decision-making? This scoping review aims to provide a panoramic portrait of the application of LLMs in CCM. Literature in seven databases, including PubMed, Embase, Scopus, Web of Science, CINAHL, IEEE Xplore, and ACM Digital Library, were searched from January 1, 2019, to June 10, 2024. Peer-reviewed journal and conference articles that discussed the application of LLMs in critical care settings were included. From an initial 619 articles, 24 were selected for final review. This review grouped applications of LLMs in CCM into three categories: clinical decision support, medical documentation and reporting, and medical education and doctor-patient communication. LLMs have advantages in handling unstructured data and do not require manual feature engineering. Meanwhile, applying LLMs to CCM faces challenges, including hallucinations, poor interpretability, bias and alignment challenges, and privacy and ethics issues. Future research should enhance model reliability and interpretability, integrate up-to-date medical knowledge, and strengthen privacy and ethical guidelines. As LLMs evolve, they could become key tools in CCM to help improve patient outcomes and optimize healthcare delivery. This study is the first review of LLMs in CCM, aiding researchers, clinicians, and policymakers to understand the current status and future potentials of LLMs in CCM.
Identifying Subgroups of ICU Patients Using End-to-End Multivariate Time-Series Clustering Algorithm Based on Real-World Vital Signs Data
Jun 03, 2023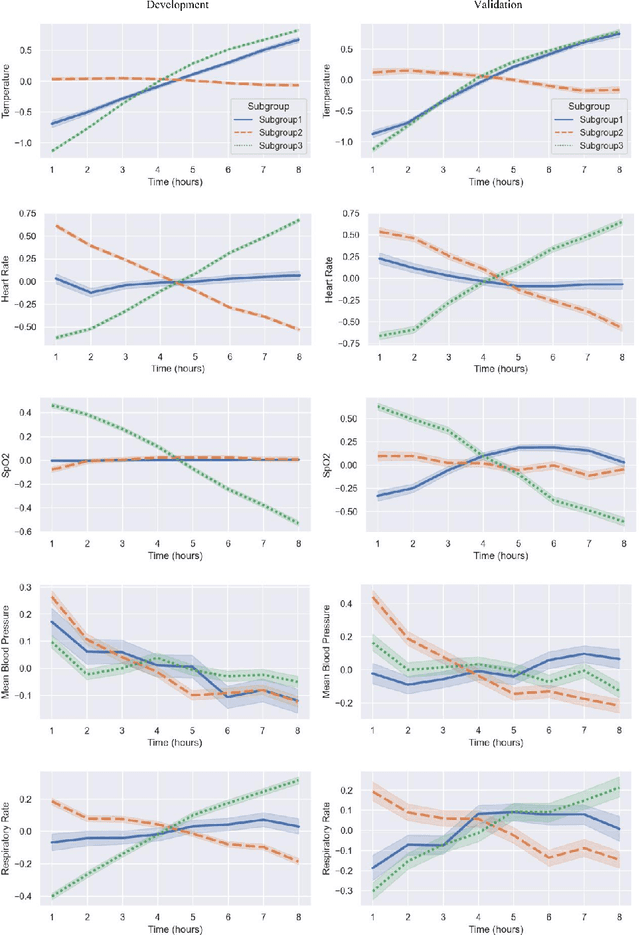
Abstract:This study employed the MIMIC-IV database as data source to investigate the use of dynamic, high-frequency, multivariate time-series vital signs data, including temperature, heart rate, mean blood pressure, respiratory rate, and SpO2, monitored first 8 hours data in the ICU stay. Various clustering algorithms were compared, and an end-to-end multivariate time series clustering system called Time2Feat, combined with K-Means, was chosen as the most effective method to cluster patients in the ICU. In clustering analysis, data of 8,080 patients admitted between 2008 and 2016 was used for model development and 2,038 patients admitted between 2017 and 2019 for model validation. By analyzing the differences in clinical mortality prognosis among different categories, varying risks of ICU mortality and hospital mortality were found between different subgroups. Furthermore, the study visualized the trajectory of vital signs changes. The findings of this study provide valuable insights into the potential use of multivariate time-series clustering systems in patient management and monitoring in the ICU setting.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge