Tijana Milenkovic
Weighted graphlets and deep neural networks for protein structure classification
Oct 07, 2019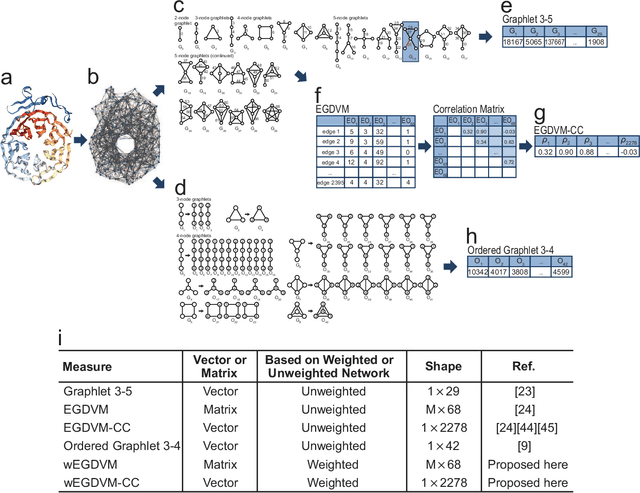


Abstract:As proteins with similar structures often have similar functions, analysis of protein structures can help predict protein functions and is thus important. We consider the problem of protein structure classification, which computationally classifies the structures of proteins into pre-defined groups. We develop a weighted network that depicts the protein structures, and more importantly, we propose the first graphlet-based measure that applies to weighted networks. Further, we develop a deep neural network (DNN) composed of both convolutional and recurrent layers to use this measure for classification. Put together, our approach shows dramatic improvements in performance over existing graphlet-based approaches on 36 real datasets. Even comparing with the state-of-the-art approach, it almost halves the classification error. In addition to protein structure networks, our weighted-graphlet measure and DNN classifier can potentially be applied to classification of other weighted networks in computational biology as well as in other domains.
The power of dynamic social networks to predict individuals' mental health
Aug 06, 2019



Abstract:Precision medicine has received attention both in and outside the clinic. We focus on the latter, by exploiting the relationship between individuals' social interactions and their mental health to develop a predictive model of one's likelihood to be depressed or anxious from rich dynamic social network data. To our knowledge, we are the first to do this. Existing studies differ from our work in at least one aspect: they do not model social interaction data as a network; they do so but analyze static network data; they examine "correlation" between social networks and health but without developing a predictive model; or they study other individual traits but not mental health. In a systematic and comprehensive evaluation, we show that our predictive model that uses dynamic social network data is superior to its static network as well as non-network equivalents when run on the same data.
Heterogeneous network approach to predict individuals' mental health
Jun 11, 2019
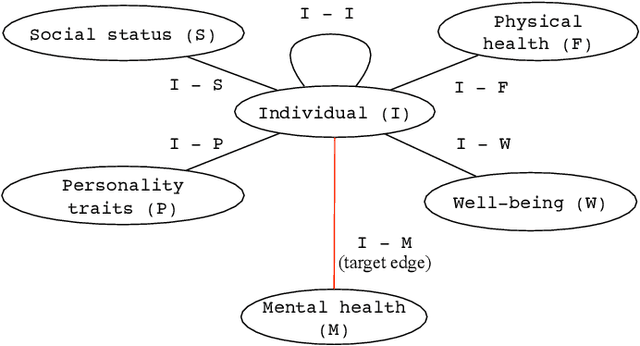


Abstract:Depression and anxiety are critical public health issues affecting millions of people around the world. To identify individuals who are vulnerable to depression and anxiety, predictive models have been built that typically utilize data from one source. Unlike these traditional models, in this study, we leverage a rich heterogeneous data set from the University of Notre Dame's NetHealth study that collected individuals' (student participants') social interaction data via smartphones, health-related behavioral data via wearables (Fitbit), and trait data from surveys. To integrate the different types of information, we model the NetHealth data as a heterogeneous information network (HIN). Then, we redefine the problem of predicting individuals' mental health conditions (depression or anxiety) in a novel manner, as applying to our HIN a popular paradigm of a recommender system (RS), which is typically used to predict the preference that a person would give to an item (e.g., a movie or book). In our case, the items are the individuals' different mental health states. We evaluate three state-of-the-art RS approaches. Also, we model the prediction of individuals' mental health as another problem type -- that of node classification (NC) in our HIN, evaluating in the process four node features under logistic regression as a proof-of-concept classifier. We find that our RS and NC network methods produce more accurate predictions than a logistic regression model using the same NetHealth data in the traditional non-network fashion as well as a random-approach. Also, we find that RS outperforms NC. This is the first study to integrate smartphone, wearable sensor, and survey data in an HIN manner and use RS or NC on the HIN to predict individuals' mental health conditions.
Network-based protein structural classification
Apr 12, 2018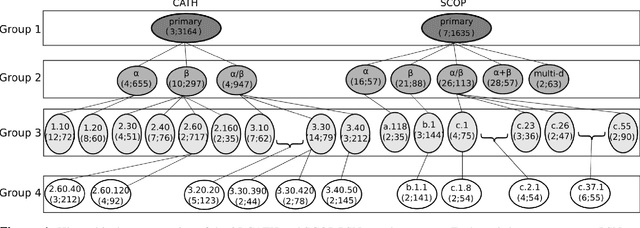
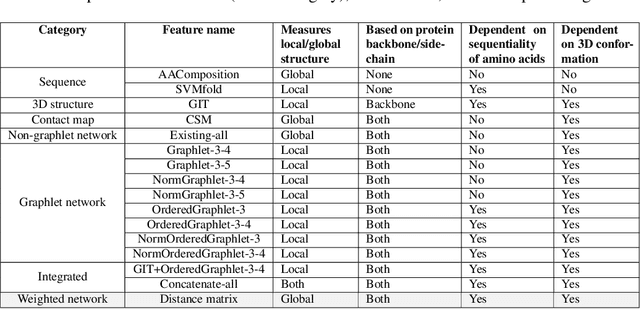
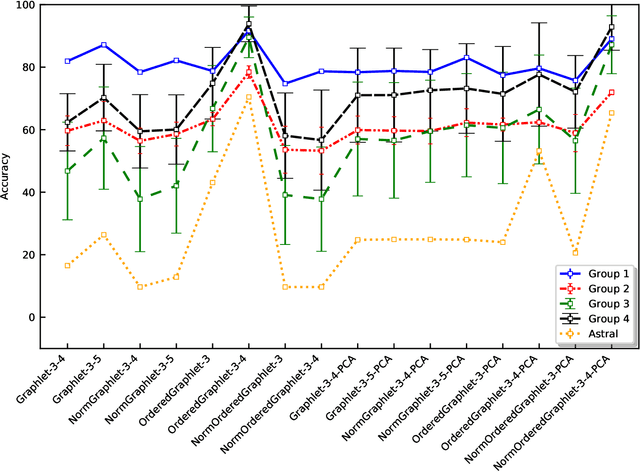
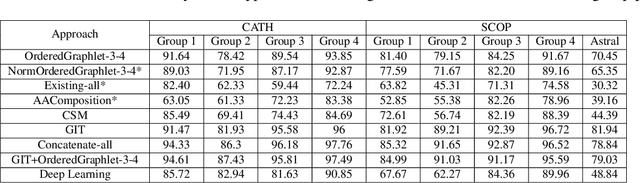
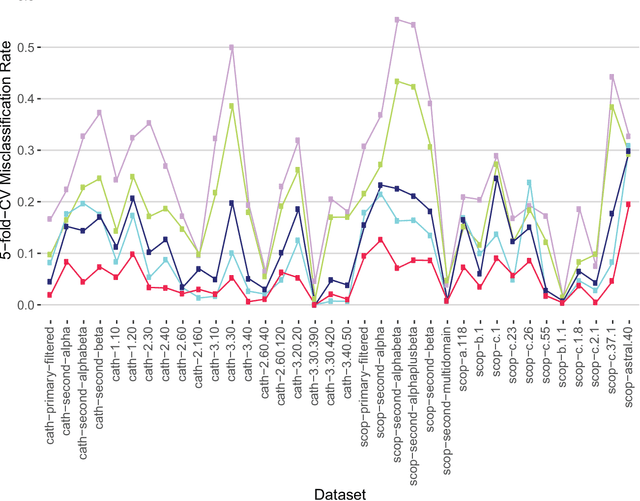
Abstract:Experimental determination of protein function is resource-consuming. As an alternative, computational prediction of protein function has received attention. In this context, protein structural classification (PSC) can help, by allowing for determining structural classes of currently unclassified proteins based on their features, and then relying on the fact that proteins with similar structures have similar functions. Existing PSC approaches rely on sequence-based or direct ("raw") 3-dimensional (3D) structure-based protein features. Instead, we first model 3D structures as protein structure networks (PSNs). Then, we use ("processed") network-based features for PSC. We are the first ones to do so. We propose the use of graphlets, state-of-the-art features in many domains of network science, in the task of PSC. Moreover, because graphlets can deal only with unweighted PSNs, and because accounting for edge weights when constructing PSNs could improve PSC accuracy, we also propose a deep learning framework that automatically learns network features from the weighted PSNs. When evaluated on a large set of 9,509 CATH and 11,451 SCOP protein domains, our proposed approaches are superior to existing PSC approaches in terms of both accuracy and running time.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge