Network-based protein structural classification
Paper and Code
Apr 12, 2018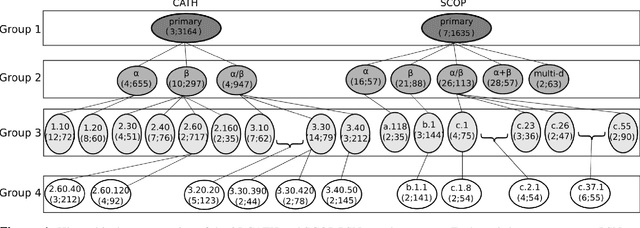
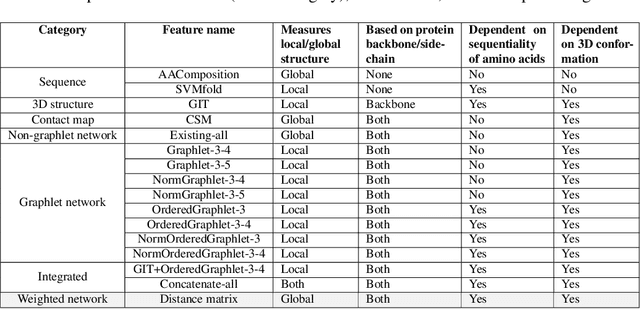
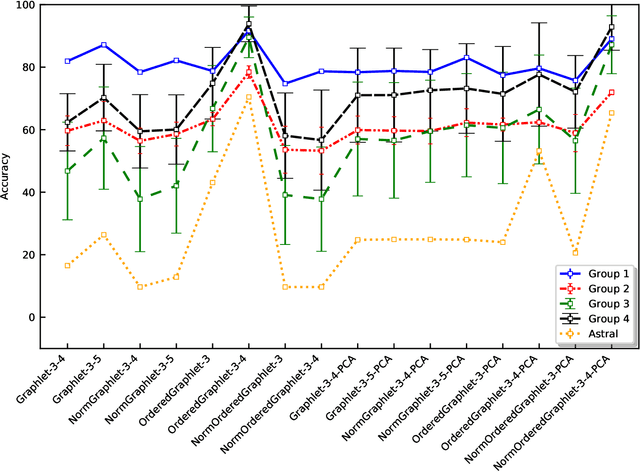
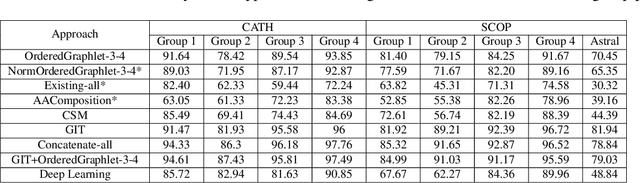
Experimental determination of protein function is resource-consuming. As an alternative, computational prediction of protein function has received attention. In this context, protein structural classification (PSC) can help, by allowing for determining structural classes of currently unclassified proteins based on their features, and then relying on the fact that proteins with similar structures have similar functions. Existing PSC approaches rely on sequence-based or direct ("raw") 3-dimensional (3D) structure-based protein features. Instead, we first model 3D structures as protein structure networks (PSNs). Then, we use ("processed") network-based features for PSC. We are the first ones to do so. We propose the use of graphlets, state-of-the-art features in many domains of network science, in the task of PSC. Moreover, because graphlets can deal only with unweighted PSNs, and because accounting for edge weights when constructing PSNs could improve PSC accuracy, we also propose a deep learning framework that automatically learns network features from the weighted PSNs. When evaluated on a large set of 9,509 CATH and 11,451 SCOP protein domains, our proposed approaches are superior to existing PSC approaches in terms of both accuracy and running time.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge