Tian Zhu
Improving Molecular Graph Generation with Flow Matching and Optimal Transport
Nov 08, 2024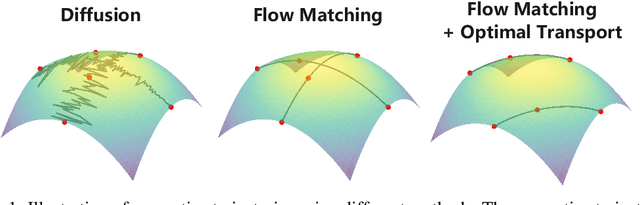

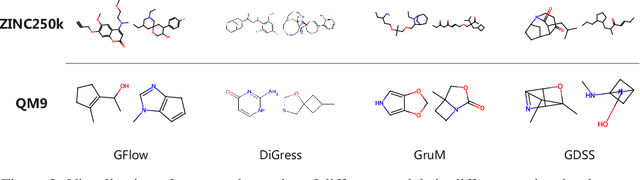

Abstract:Generating molecular graphs is crucial in drug design and discovery but remains challenging due to the complex interdependencies between nodes and edges. While diffusion models have demonstrated their potentiality in molecular graph design, they often suffer from unstable training and inefficient sampling. To enhance generation performance and training stability, we propose GGFlow, a discrete flow matching generative model incorporating optimal transport for molecular graphs and it incorporates an edge-augmented graph transformer to enable the direct communications among chemical bounds. Additionally, GGFlow introduces a novel goal-guided generation framework to control the generative trajectory of our model, aiming to design novel molecular structures with the desired properties. GGFlow demonstrates superior performance on both unconditional and conditional molecule generation tasks, outperforming existing baselines and underscoring its effectiveness and potential for wider application.
Predicting mutational effects on protein-protein binding via a side-chain diffusion probabilistic model
Oct 30, 2023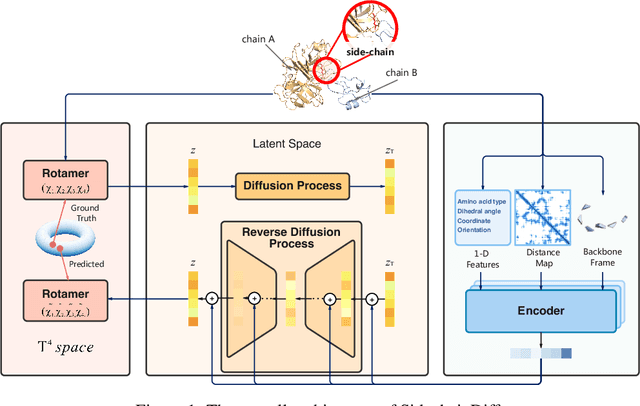
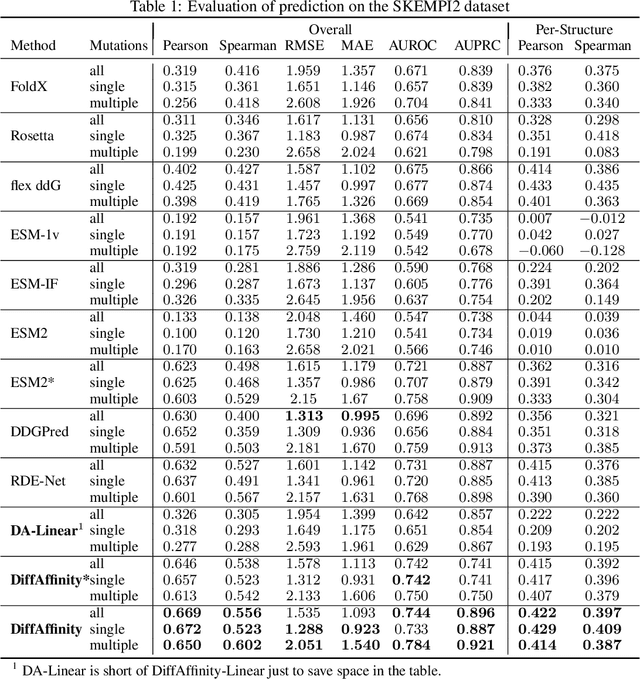
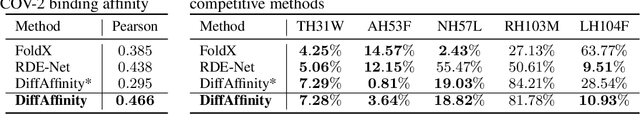
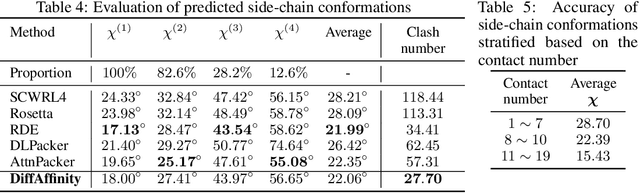
Abstract:Many crucial biological processes rely on networks of protein-protein interactions. Predicting the effect of amino acid mutations on protein-protein binding is vital in protein engineering and therapeutic discovery. However, the scarcity of annotated experimental data on binding energy poses a significant challenge for developing computational approaches, particularly deep learning-based methods. In this work, we propose SidechainDiff, a representation learning-based approach that leverages unlabelled experimental protein structures. SidechainDiff utilizes a Riemannian diffusion model to learn the generative process of side-chain conformations and can also give the structural context representations of mutations on the protein-protein interface. Leveraging the learned representations, we achieve state-of-the-art performance in predicting the mutational effects on protein-protein binding. Furthermore, SidechainDiff is the first diffusion-based generative model for side-chains, distinguishing it from prior efforts that have predominantly focused on generating protein backbone structures.
Opinion Extraction as A Structured Sentiment Analysis using Transformers
Dec 09, 2021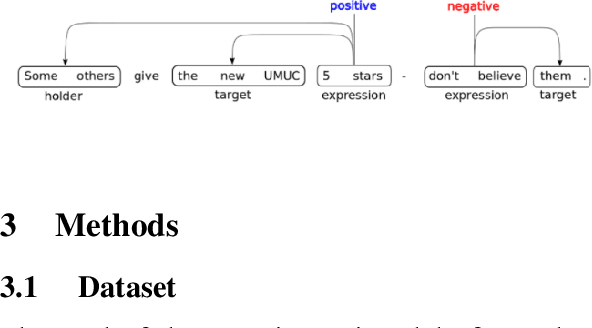
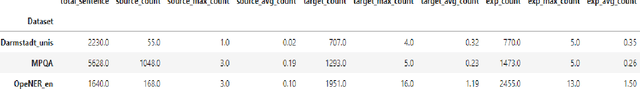
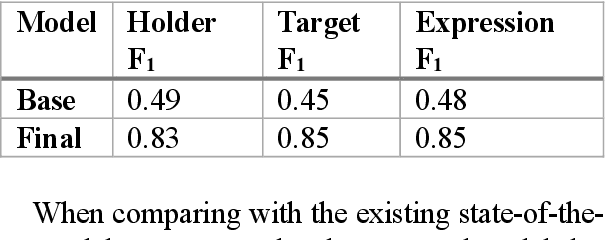
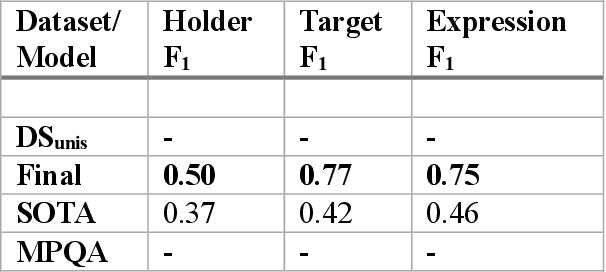
Abstract:Relationship extraction and named entity recognition have always been considered as two distinct tasks that require different input data, labels, and models. However, both are essential for structured sentiment analysis. We believe that both tasks can be combined into a single stacked model with the same input data. We performed different experiments to find the best model to extract multiple opinion tuples from a single sentence. The opinion tuples will consist of holders, targets, and expressions. With the opinion tuples, we will be able to extract the relationship we need.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge