Thomas C. Booth
Autonomous navigation of catheters and guidewires in mechanical thrombectomy using inverse reinforcement learning
Jun 18, 2024Abstract:Purpose: Autonomous navigation of catheters and guidewires can enhance endovascular surgery safety and efficacy, reducing procedure times and operator radiation exposure. Integrating tele-operated robotics could widen access to time-sensitive emergency procedures like mechanical thrombectomy (MT). Reinforcement learning (RL) shows potential in endovascular navigation, yet its application encounters challenges without a reward signal. This study explores the viability of autonomous navigation in MT vasculature using inverse RL (IRL) to leverage expert demonstrations. Methods: This study established a simulation-based training and evaluation environment for MT navigation. We used IRL to infer reward functions from expert behaviour when navigating a guidewire and catheter. We utilized soft actor-critic to train models with various reward functions and compared their performance in silico. Results: We demonstrated feasibility of navigation using IRL. When evaluating single versus dual device (i.e. guidewire versus catheter and guidewire) tracking, both methods achieved high success rates of 95% and 96%, respectively. Dual-tracking, however, utilized both devices mimicking an expert. A success rate of 100% and procedure time of 22.6 s were obtained when training with a reward function obtained through reward shaping. This outperformed a dense reward function (96%, 24.9 s) and an IRL-derived reward function (48%, 59.2 s). Conclusions: We have contributed to the advancement of autonomous endovascular intervention navigation, particularly MT, by employing IRL. The results underscore the potential of using reward shaping to train models, offering a promising avenue for enhancing the accessibility and precision of MT. We envisage that future research can extend our methodology to diverse anatomical structures to enhance generalizability.
* Abstract shortened for arXiv character limit
A self-supervised text-vision framework for automated brain abnormality detection
May 05, 2024



Abstract:Artificial neural networks trained on large, expert-labelled datasets are considered state-of-the-art for a range of medical image recognition tasks. However, categorically labelled datasets are time-consuming to generate and constrain classification to a pre-defined, fixed set of classes. For neuroradiological applications in particular, this represents a barrier to clinical adoption. To address these challenges, we present a self-supervised text-vision framework that learns to detect clinically relevant abnormalities in brain MRI scans by directly leveraging the rich information contained in accompanying free-text neuroradiology reports. Our training approach consisted of two-steps. First, a dedicated neuroradiological language model - NeuroBERT - was trained to generate fixed-dimensional vector representations of neuroradiology reports (N = 50,523) via domain-specific self-supervised learning tasks. Next, convolutional neural networks (one per MRI sequence) learnt to map individual brain scans to their corresponding text vector representations by optimising a mean square error loss. Once trained, our text-vision framework can be used to detect abnormalities in unreported brain MRI examinations by scoring scans against suitable query sentences (e.g., 'there is an acute stroke', 'there is hydrocephalus' etc.), enabling a range of classification-based applications including automated triage. Potentially, our framework could also serve as a clinical decision support tool, not only by suggesting findings to radiologists and detecting errors in provisional reports, but also by retrieving and displaying examples of pathologies from historical examinations that could be relevant to the current case based on textual descriptors.
Automated triaging of head MRI examinations using convolutional neural networks
Jun 15, 2021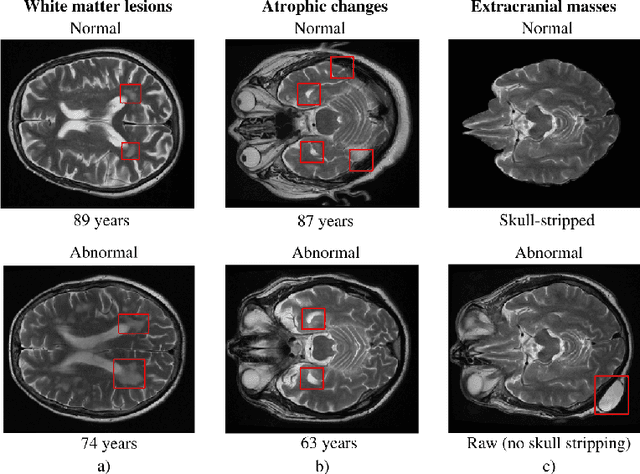
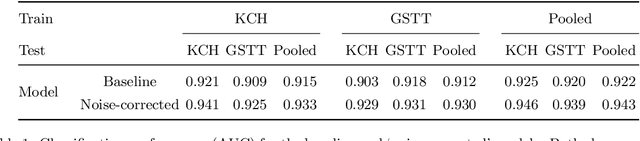
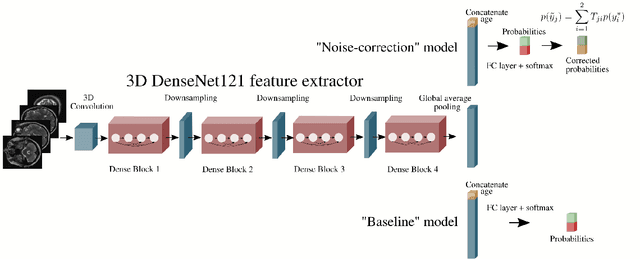
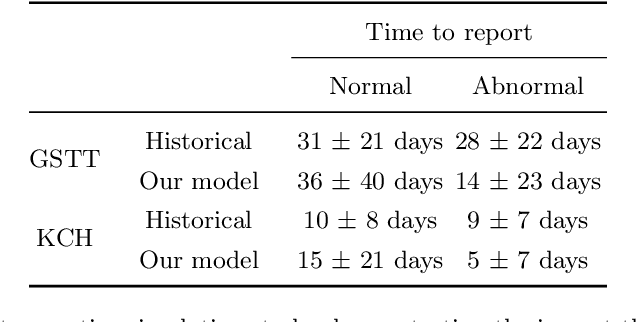
Abstract:The growing demand for head magnetic resonance imaging (MRI) examinations, along with a global shortage of radiologists, has led to an increase in the time taken to report head MRI scans around the world. For many neurological conditions, this delay can result in increased morbidity and mortality. An automated triaging tool could reduce reporting times for abnormal examinations by identifying abnormalities at the time of imaging and prioritizing the reporting of these scans. In this work, we present a convolutional neural network for detecting clinically-relevant abnormalities in $\text{T}_2$-weighted head MRI scans. Using a validated neuroradiology report classifier, we generated a labelled dataset of 43,754 scans from two large UK hospitals for model training, and demonstrate accurate classification (area under the receiver operating curve (AUC) = 0.943) on a test set of 800 scans labelled by a team of neuroradiologists. Importantly, when trained on scans from only a single hospital the model generalized to scans from the other hospital ($\Delta$AUC $\leq$ 0.02). A simulation study demonstrated that our model would reduce the mean reporting time for abnormal examinations from 28 days to 14 days and from 9 days to 5 days at the two hospitals, demonstrating feasibility for use in a clinical triage environment.
Labelling imaging datasets on the basis of neuroradiology reports: a validation study
Jul 08, 2020

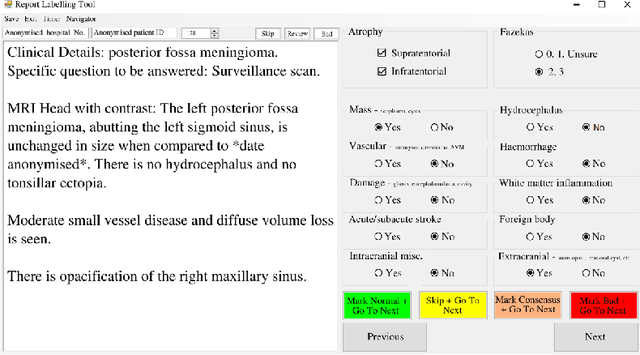
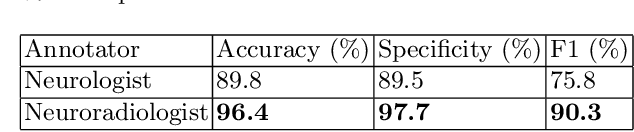
Abstract:Natural language processing (NLP) shows promise as a means to automate the labelling of hospital-scale neuroradiology magnetic resonance imaging (MRI) datasets for computer vision applications. To date, however, there has been no thorough investigation into the validity of this approach, including determining the accuracy of report labels compared to image labels as well as examining the performance of non-specialist labellers. In this work, we draw on the experience of a team of neuroradiologists who labelled over 5000 MRI neuroradiology reports as part of a project to build a dedicated deep learning-based neuroradiology report classifier. We show that, in our experience, assigning binary labels (i.e. normal vs abnormal) to images from reports alone is highly accurate. In contrast to the binary labels, however, the accuracy of more granular labelling is dependent on the category, and we highlight reasons for this discrepancy. We also show that downstream model performance is reduced when labelling of training reports is performed by a non-specialist. To allow other researchers to accelerate their research, we make our refined abnormality definitions and labelling rules available, as well as our easy-to-use radiology report labelling app which helps streamline this process.
Automated Labelling using an Attention model for Radiology reports of MRI scans (ALARM)
Feb 16, 2020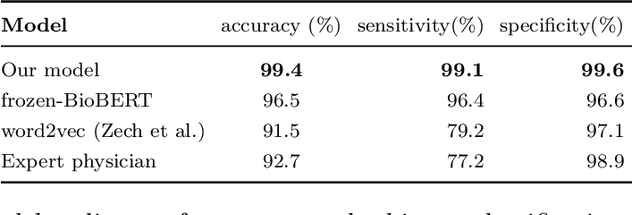
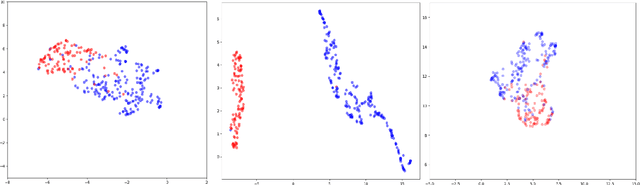
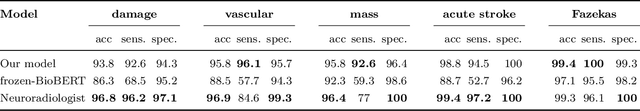
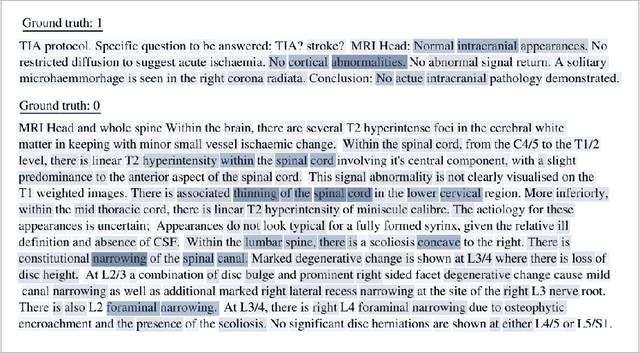
Abstract:Labelling large datasets for training high-capacity neural networks is a major obstacle to the development of deep learning-based medical imaging applications. Here we present a transformer-based network for magnetic resonance imaging (MRI) radiology report classification which automates this task by assigning image labels on the basis of free-text expert radiology reports. Our model's performance is comparable to that of an expert radiologist, and better than that of an expert physician, demonstrating the feasibility of this approach. We make code available online for researchers to label their own MRI datasets for medical imaging applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge