Theresa Willem
AI Alignment in Medical Imaging: Unveiling Hidden Biases Through Counterfactual Analysis
Apr 28, 2025Abstract:Machine learning (ML) systems for medical imaging have demonstrated remarkable diagnostic capabilities, but their susceptibility to biases poses significant risks, since biases may negatively impact generalization performance. In this paper, we introduce a novel statistical framework to evaluate the dependency of medical imaging ML models on sensitive attributes, such as demographics. Our method leverages the concept of counterfactual invariance, measuring the extent to which a model's predictions remain unchanged under hypothetical changes to sensitive attributes. We present a practical algorithm that combines conditional latent diffusion models with statistical hypothesis testing to identify and quantify such biases without requiring direct access to counterfactual data. Through experiments on synthetic datasets and large-scale real-world medical imaging datasets, including \textsc{cheXpert} and MIMIC-CXR, we demonstrate that our approach aligns closely with counterfactual fairness principles and outperforms standard baselines. This work provides a robust tool to ensure that ML diagnostic systems generalize well, e.g., across demographic groups, offering a critical step towards AI safety in healthcare. Code: https://github.com/Neferpitou3871/AI-Alignment-Medical-Imaging.
Attention-based Saliency Maps Improve Interpretability of Pneumothorax Classification
Mar 03, 2023



Abstract:Purpose: To investigate chest radiograph (CXR) classification performance of vision transformers (ViT) and interpretability of attention-based saliency using the example of pneumothorax classification. Materials and Methods: In this retrospective study, ViTs were fine-tuned for lung disease classification using four public data sets: CheXpert, Chest X-Ray 14, MIMIC CXR, and VinBigData. Saliency maps were generated using transformer multimodal explainability and gradient-weighted class activation mapping (GradCAM). Classification performance was evaluated on the Chest X-Ray 14, VinBigData, and SIIM-ACR data sets using the area under the receiver operating characteristic curve analysis (AUC) and compared with convolutional neural networks (CNNs). The explainability methods were evaluated with positive/negative perturbation, sensitivity-n, effective heat ratio, intra-architecture repeatability and interarchitecture reproducibility. In the user study, three radiologists classified 160 CXRs with/without saliency maps for pneumothorax and rated their usefulness. Results: ViTs had comparable CXR classification AUCs compared with state-of-the-art CNNs 0.95 (95% CI: 0.943, 0.950) versus 0.83 (95%, CI 0.826, 0.842) on Chest X-Ray 14, 0.84 (95% CI: 0.769, 0.912) versus 0.83 (95% CI: 0.760, 0.895) on VinBigData, and 0.85 (95% CI: 0.847, 0.861) versus 0.87 (95% CI: 0.868, 0.882) on SIIM ACR. Both saliency map methods unveiled a strong bias toward pneumothorax tubes in the models. Radiologists found 47% of the attention-based saliency maps useful and 39% of GradCAM. The attention-based methods outperformed GradCAM on all metrics. Conclusion: ViTs performed similarly to CNNs in CXR classification, and their attention-based saliency maps were more useful to radiologists and outperformed GradCAM.
A knee cannot have lung disease: out-of-distribution detection with in-distribution voting using the medical example of chest X-ray classification
Aug 01, 2022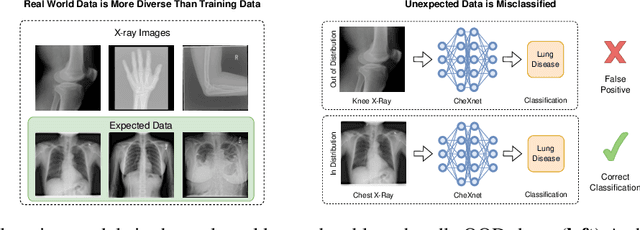
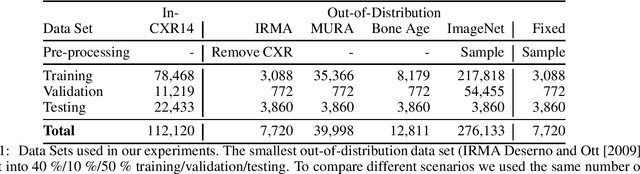
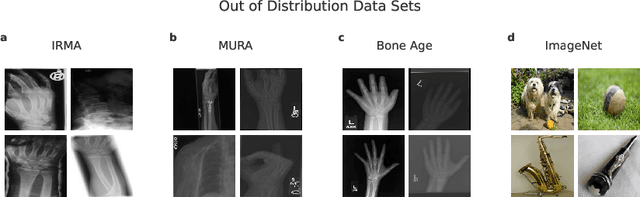
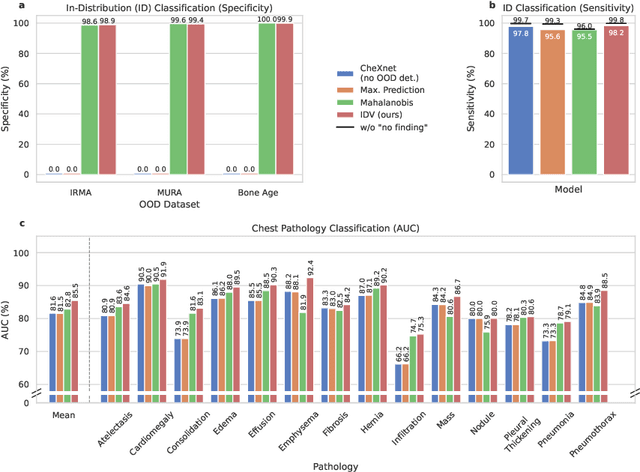
Abstract:Deep learning models are being applied to more and more use cases with astonishing success stories, but how do they perform in the real world? To test a model, a specific cleaned data set is assembled. However, when deployed in the real world, the model will face unexpected, out-of-distribution (OOD) data. In this work, we show that the so-called "radiologist-level" CheXnet model fails to recognize all OOD images and classifies them as having lung disease. To address this issue, we propose in-distribution voting, a novel method to classify out-of-distribution images for multi-label classification. Using independent class-wise in-distribution (ID) predictors trained on ID and OOD data we achieve, on average, 99 % ID classification specificity and 98 % sensitivity, improving the end-to-end performance significantly compared to previous works on the chest X-ray 14 data set. Our method surpasses other output-based OOD detectors even when trained solely with ImageNet as OOD data and tested with X-ray OOD images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge