Alessandro Wollek
Exploring the Impact of Image Resolution on Chest X-ray Classification Performance
Jun 09, 2023



Abstract:Deep learning models for image classification have often used a resolution of $224\times224$ pixels for computational reasons. This study investigates the effect of image resolution on chest X-ray classification performance, using the ChestX-ray14 dataset. The results show that a higher image resolution, specifically $1024\times1024$ pixels, has the best overall classification performance, with a slight decline in performance between $256\times256$ to $512\times512$ pixels for most of the pathological classes. Comparison of saliency map-generated bounding boxes revealed that commonly used resolutions are insufficient for finding most pathologies.
Automated Labeling of German Chest X-Ray Radiology Reports using Deep Learning
Jun 09, 2023



Abstract:Radiologists are in short supply globally, and deep learning models offer a promising solution to address this shortage as part of clinical decision-support systems. However, training such models often requires expensive and time-consuming manual labeling of large datasets. Automatic label extraction from radiology reports can reduce the time required to obtain labeled datasets, but this task is challenging due to semantically similar words and missing annotated data. In this work, we explore the potential of weak supervision of a deep learning-based label prediction model, using a rule-based labeler. We propose a deep learning-based CheXpert label prediction model, pre-trained on reports labeled by a rule-based German CheXpert model and fine-tuned on a small dataset of manually labeled reports. Our results demonstrate the effectiveness of our approach, which significantly outperformed the rule-based model on all three tasks. Our findings highlight the benefits of employing deep learning-based models even in scenarios with sparse data and the use of the rule-based labeler as a tool for weak supervision.
WindowNet: Learnable Windows for Chest X-ray Classification
Jun 09, 2023



Abstract:Chest X-ray (CXR) images are commonly compressed to a lower resolution and bit depth to reduce their size, potentially altering subtle diagnostic features. Radiologists use windowing operations to enhance image contrast, but the impact of such operations on CXR classification performance is unclear. In this study, we show that windowing can improve CXR classification performance, and propose WindowNet, a model that learns optimal window settings. We first investigate the impact of bit-depth on classification performance and find that a higher bit-depth (12-bit) leads to improved performance. We then evaluate different windowing settings and show that training with a distinct window generally improves pathology-wise classification performance. Finally, we propose and evaluate WindowNet, a model that learns optimal window settings, and show that it significantly improves performance compared to the baseline model without windowing.
German CheXpert Chest X-ray Radiology Report Labeler
Jun 05, 2023Abstract:This study aimed to develop an algorithm to automatically extract annotations for chest X-ray classification models from German thoracic radiology reports. An automatic label extraction model was designed based on the CheXpert architecture, and a web-based annotation interface was created for iterative improvements. Results showed that automated label extraction can reduce time spent on manual labeling and improve overall modeling performance. The model trained on automatically extracted labels performed competitively to manually labeled data and strongly outperformed the model trained on publicly available data.
Attention-based Saliency Maps Improve Interpretability of Pneumothorax Classification
Mar 03, 2023



Abstract:Purpose: To investigate chest radiograph (CXR) classification performance of vision transformers (ViT) and interpretability of attention-based saliency using the example of pneumothorax classification. Materials and Methods: In this retrospective study, ViTs were fine-tuned for lung disease classification using four public data sets: CheXpert, Chest X-Ray 14, MIMIC CXR, and VinBigData. Saliency maps were generated using transformer multimodal explainability and gradient-weighted class activation mapping (GradCAM). Classification performance was evaluated on the Chest X-Ray 14, VinBigData, and SIIM-ACR data sets using the area under the receiver operating characteristic curve analysis (AUC) and compared with convolutional neural networks (CNNs). The explainability methods were evaluated with positive/negative perturbation, sensitivity-n, effective heat ratio, intra-architecture repeatability and interarchitecture reproducibility. In the user study, three radiologists classified 160 CXRs with/without saliency maps for pneumothorax and rated their usefulness. Results: ViTs had comparable CXR classification AUCs compared with state-of-the-art CNNs 0.95 (95% CI: 0.943, 0.950) versus 0.83 (95%, CI 0.826, 0.842) on Chest X-Ray 14, 0.84 (95% CI: 0.769, 0.912) versus 0.83 (95% CI: 0.760, 0.895) on VinBigData, and 0.85 (95% CI: 0.847, 0.861) versus 0.87 (95% CI: 0.868, 0.882) on SIIM ACR. Both saliency map methods unveiled a strong bias toward pneumothorax tubes in the models. Radiologists found 47% of the attention-based saliency maps useful and 39% of GradCAM. The attention-based methods outperformed GradCAM on all metrics. Conclusion: ViTs performed similarly to CNNs in CXR classification, and their attention-based saliency maps were more useful to radiologists and outperformed GradCAM.
A knee cannot have lung disease: out-of-distribution detection with in-distribution voting using the medical example of chest X-ray classification
Aug 01, 2022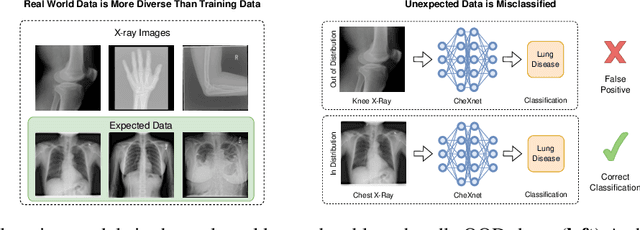
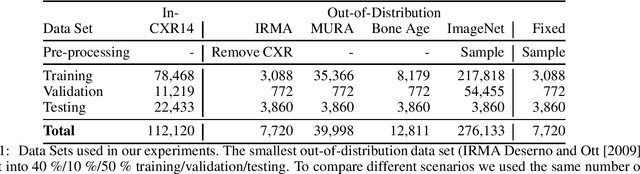
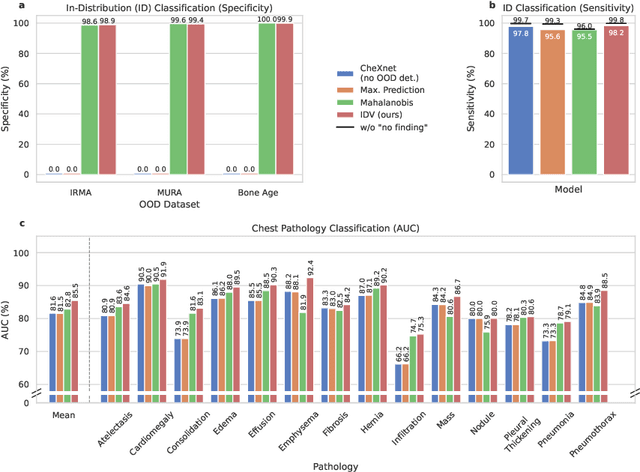
Abstract:Deep learning models are being applied to more and more use cases with astonishing success stories, but how do they perform in the real world? To test a model, a specific cleaned data set is assembled. However, when deployed in the real world, the model will face unexpected, out-of-distribution (OOD) data. In this work, we show that the so-called "radiologist-level" CheXnet model fails to recognize all OOD images and classifies them as having lung disease. To address this issue, we propose in-distribution voting, a novel method to classify out-of-distribution images for multi-label classification. Using independent class-wise in-distribution (ID) predictors trained on ID and OOD data we achieve, on average, 99 % ID classification specificity and 98 % sensitivity, improving the end-to-end performance significantly compared to previous works on the chest X-ray 14 data set. Our method surpasses other output-based OOD detectors even when trained solely with ImageNet as OOD data and tested with X-ray OOD images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge