Theodoros Theodoridis
Deriving Hematological Disease Classes Using Fuzzy Logic and Expert Knowledge: A Comprehensive Machine Learning Approach with CBC Parameters
Jun 18, 2024Abstract:In the intricate field of medical diagnostics, capturing the subtle manifestations of diseases remains a challenge. Traditional methods, often binary in nature, may not encapsulate the nuanced variances that exist in real-world clinical scenarios. This paper introduces a novel approach by leveraging Fuzzy Logic Rules to derive disease classes based on expert domain knowledge from a medical practitioner. By recognizing that diseases do not always fit into neat categories, and that expert knowledge can guide the fuzzification of these boundaries, our methodology offers a more sophisticated and nuanced diagnostic tool. Using a dataset procured from a prominent hospital, containing detailed patient blood count records, we harness Fuzzy Logic Rules, a computational technique celebrated for its ability to handle ambiguity. This approach, moving through stages of fuzzification, rule application, inference, and ultimately defuzzification, produces refined diagnostic predictions. When combined with the Random Forest classifier, the system adeptly predicts hematological conditions using Complete Blood Count (CBC) parameters. Preliminary results showcase high accuracy levels, underscoring the advantages of integrating fuzzy logic into the diagnostic process. When juxtaposed with traditional diagnostic techniques, it becomes evident that Fuzzy Logic, especially when guided by medical expertise, offers significant advancements in the realm of hematological diagnostics. This paper not only paves the path for enhanced patient care but also beckons a deeper dive into the potentialities of fuzzy logic in various medical diagnostic applications.
Tensor Comprehensions: Framework-Agnostic High-Performance Machine Learning Abstractions
Jun 29, 2018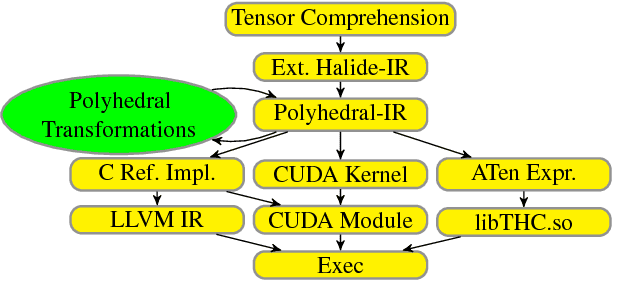
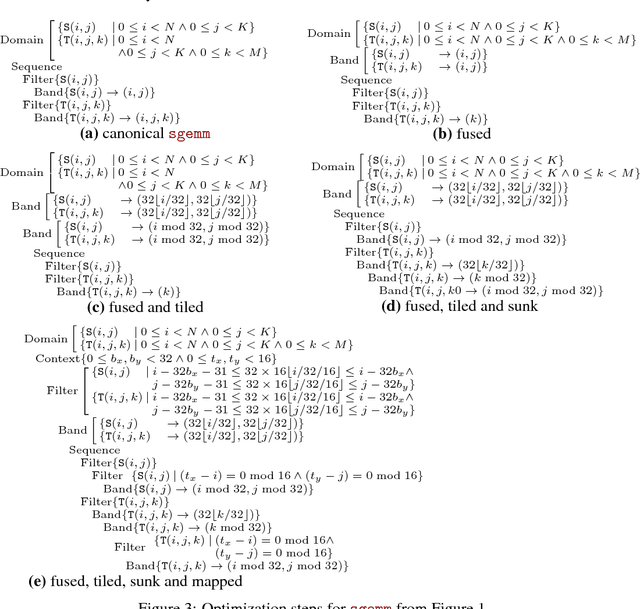
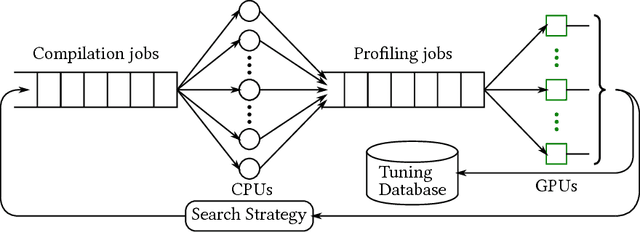
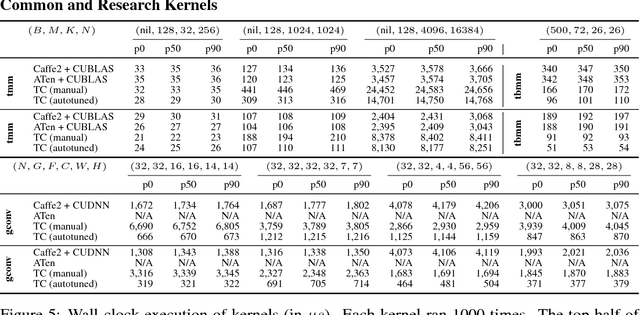
Abstract:Deep learning models with convolutional and recurrent networks are now ubiquitous and analyze massive amounts of audio, image, video, text and graph data, with applications in automatic translation, speech-to-text, scene understanding, ranking user preferences, ad placement, etc. Competing frameworks for building these networks such as TensorFlow, Chainer, CNTK, Torch/PyTorch, Caffe1/2, MXNet and Theano, explore different tradeoffs between usability and expressiveness, research or production orientation and supported hardware. They operate on a DAG of computational operators, wrapping high-performance libraries such as CUDNN (for NVIDIA GPUs) or NNPACK (for various CPUs), and automate memory allocation, synchronization, distribution. Custom operators are needed where the computation does not fit existing high-performance library calls, usually at a high engineering cost. This is frequently required when new operators are invented by researchers: such operators suffer a severe performance penalty, which limits the pace of innovation. Furthermore, even if there is an existing runtime call these frameworks can use, it often doesn't offer optimal performance for a user's particular network architecture and dataset, missing optimizations between operators as well as optimizations that can be done knowing the size and shape of data. Our contributions include (1) a language close to the mathematics of deep learning called Tensor Comprehensions, (2) a polyhedral Just-In-Time compiler to convert a mathematical description of a deep learning DAG into a CUDA kernel with delegated memory management and synchronization, also providing optimizations such as operator fusion and specialization for specific sizes, (3) a compilation cache populated by an autotuner. [Abstract cutoff]
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge