Taylor L. Bobrow
Multi-contrast laser endoscopy for in vivo gastrointestinal imaging
May 15, 2025



Abstract:White light endoscopy is the clinical gold standard for detecting diseases in the gastrointestinal tract. Most applications involve identifying visual abnormalities in tissue color, texture, and shape. Unfortunately, the contrast of these features is often subtle, causing many clinically relevant cases to go undetected. To overcome this challenge, we introduce Multi-contrast Laser Endoscopy (MLE): a platform for widefield clinical imaging with rapidly tunable spectral, coherent, and directional illumination. We demonstrate three capabilities of MLE: enhancing tissue chromophore contrast with multispectral diffuse reflectance, quantifying blood flow using laser speckle contrast imaging, and characterizing mucosal topography using photometric stereo. We validate MLE with benchtop models, then demonstrate MLE in vivo during clinical colonoscopies. MLE images from 31 polyps demonstrate an approximate three-fold improvement in contrast and a five-fold improvement in color difference compared to white light and narrow band imaging. With the ability to reveal multiple complementary types of tissue contrast while seamlessly integrating into the clinical environment, MLE shows promise as an investigative tool to improve gastrointestinal imaging.
Colonoscopy 3D Video Dataset with Paired Depth from 2D-3D Registration
Jun 17, 2022



Abstract:Screening colonoscopy is an important clinical application for several 3D computer vision techniques, including depth estimation, surface reconstruction, and missing region detection. However, the development, evaluation, and comparison of these techniques in real colonoscopy videos remain largely qualitative due to the difficulty of acquiring ground truth data. In this work, we present a Colonoscopy 3D Video Dataset (C3VD) acquired with a high definition clinical colonoscope and high-fidelity colon models for benchmarking computer vision methods in colonoscopy. We introduce a novel multimodal 2D-3D registration technique to register optical video sequences with ground truth rendered views of a known 3D model. The different modalities are registered by transforming optical images to depth maps with a Generative Adversarial Network and aligning edge features with an evolutionary optimizer. This registration method achieves an average translation error of 0.321 millimeters and an average rotation error of 0.159 degrees in simulation experiments where error-free ground truth is available. The method also leverages video information, improving registration accuracy by 55.6% for translation and 60.4% for rotation compared to single frame registration. 22 short video sequences were registered to generate 10,015 total frames with paired ground truth depth, surface normals, optical flow, occlusion, six degree-of-freedom pose, coverage maps, and 3D models. The dataset also includes screening videos acquired by a gastroenterologist with paired ground truth pose and 3D surface models. The dataset and registration source code are available at durr.jhu.edu/C3VD.
GAN Inversion for Data Augmentation to Improve Colonoscopy Lesion Classification
May 04, 2022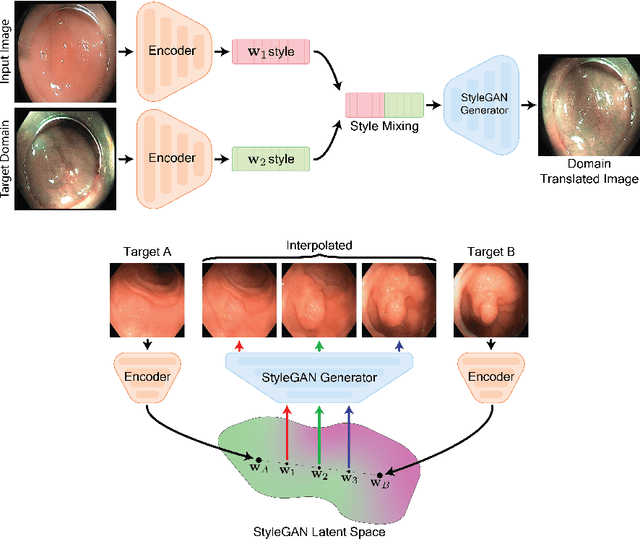

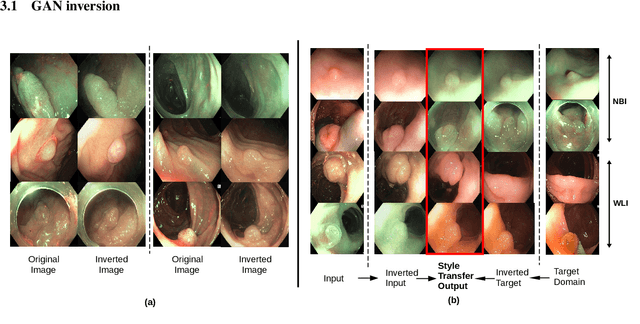
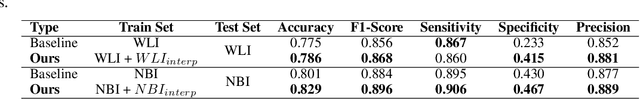
Abstract:A major challenge in applying deep learning to medical imaging is the paucity of annotated data. This study demonstrates that synthetic colonoscopy images generated by Generative Adversarial Network (GAN) inversion can be used as training data to improve the lesion classification performance of deep learning models. This approach inverts pairs of images with the same label to a semantically rich & disentangled latent space and manipulates latent representations to produce new synthetic images with the same label. We perform image modality translation (style transfer) between white light and narrowband imaging (NBI). We also generate realistic-looking synthetic lesion images by interpolating between original training images to increase the variety of lesion shapes in the training dataset. We show that these approaches outperform comparative colonoscopy data augmentation techniques without the need to re-train multiple generative models. This approach also leverages information from datasets that may not have been designed for the specific colonoscopy downstream task. E.g. using a bowel prep grading dataset for a polyp classification task. Our experiments show this approach can perform multiple colonoscopy data augmentations, which improve the downstream polyp classification performance over baseline and comparison methods by up to 6%.
Improving colonoscopy lesion classification using semi-supervised deep learning
Sep 07, 2020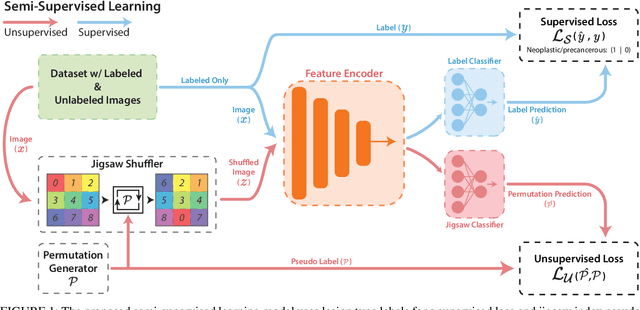

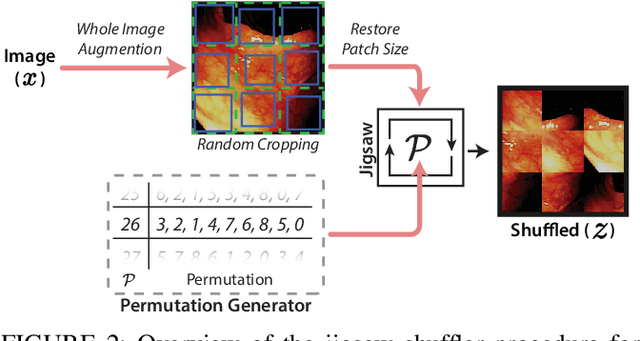
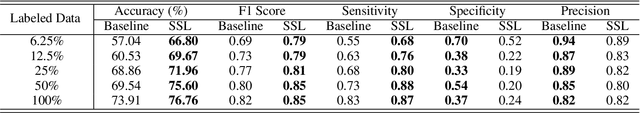
Abstract:While data-driven approaches excel at many image analysis tasks, the performance of these approaches is often limited by a shortage of annotated data available for training. Recent work in semi-supervised learning has shown that meaningful representations of images can be obtained from training with large quantities of unlabeled data, and that these representations can improve the performance of supervised tasks. Here, we demonstrate that an unsupervised jigsaw learning task, in combination with supervised training, results in up to a 9.8% improvement in correctly classifying lesions in colonoscopy images when compared to a fully-supervised baseline. We additionally benchmark improvements in domain adaptation and out-of-distribution detection, and demonstrate that semi-supervised learning outperforms supervised learning in both cases. In colonoscopy applications, these metrics are important given the skill required for endoscopic assessment of lesions, the wide variety of endoscopy systems in use, and the homogeneity that is typical of labeled datasets.
SLAM Endoscopy enhanced by adversarial depth prediction
Jun 29, 2019



Abstract:Medical endoscopy remains a challenging application for simultaneous localization and mapping (SLAM) due to the sparsity of image features and size constraints that prevent direct depth-sensing. We present a SLAM approach that incorporates depth predictions made by an adversarially-trained convolutional neural network (CNN) applied to monocular endoscopy images. The depth network is trained with synthetic images of a simple colon model, and then fine-tuned with domain-randomized, photorealistic images rendered from computed tomography measurements of human colons. Each image is paired with an error-free depth map for supervised adversarial learning. Monocular RGB images are then fused with corresponding depth predictions, enabling dense reconstruction and mosaicing as an endoscope is advanced through the gastrointestinal tract. Our preliminary results demonstrate that incorporating monocular depth estimation into a SLAM architecture can enable dense reconstruction of endoscopic scenes.
DeepLSR: Deep learning approach for laser speckle reduction
Oct 23, 2018



Abstract:We present a deep learning approach for laser speckle reduction ('DeepLSR') on images illuminated with a multi-wavelength, red-green-blue laser. We acquired a set of images from a variety of objects illuminated with laser light, both with and without optical speckle reduction, and an incoherent light-emitting diode. An adversarial network was then trained for paired image-to-image translation to transform images from a source domain of coherent illumination to a target domain of incoherent illumination. When applied to a new image set of coherently-illuminated test objects, this network reconstructs incoherently-illuminated images with an average peak signal-to-noise ratio and structural similarity index of 36 dB and 0.91, respectively, compared to 30 dB and 0.88 using optical speckle reduction, and 30 dB and 0.88 using non-local means processing. We demonstrate proof-of-concept for speckle-reduced laser endoscopy by applying DeepLSR to images of ex-vivo gastrointestinal tissue illuminated with a fiber-coupled laser source. For applications that require speckle-reduced imaging, DeepLSR holds promise to enable the use of coherent sources that are more stable, efficient, compact, and brighter than conventional alternatives.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge