GAN Inversion for Data Augmentation to Improve Colonoscopy Lesion Classification
Paper and Code
May 04, 2022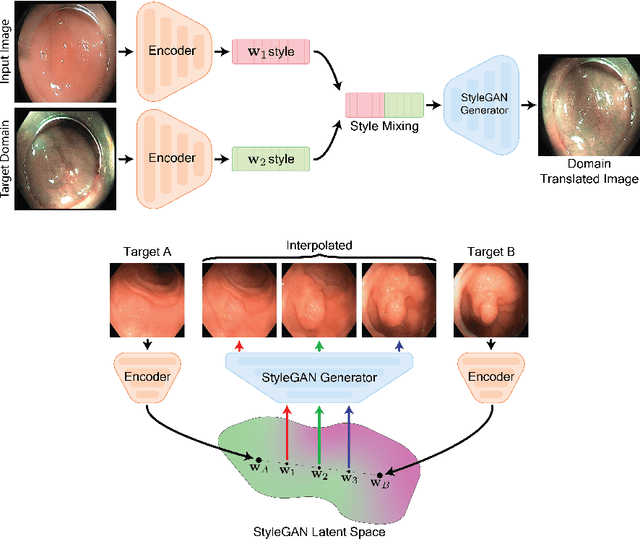

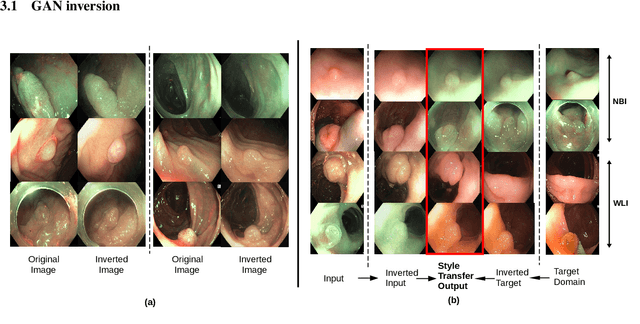
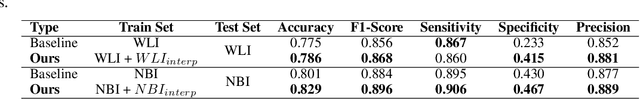
A major challenge in applying deep learning to medical imaging is the paucity of annotated data. This study demonstrates that synthetic colonoscopy images generated by Generative Adversarial Network (GAN) inversion can be used as training data to improve the lesion classification performance of deep learning models. This approach inverts pairs of images with the same label to a semantically rich & disentangled latent space and manipulates latent representations to produce new synthetic images with the same label. We perform image modality translation (style transfer) between white light and narrowband imaging (NBI). We also generate realistic-looking synthetic lesion images by interpolating between original training images to increase the variety of lesion shapes in the training dataset. We show that these approaches outperform comparative colonoscopy data augmentation techniques without the need to re-train multiple generative models. This approach also leverages information from datasets that may not have been designed for the specific colonoscopy downstream task. E.g. using a bowel prep grading dataset for a polyp classification task. Our experiments show this approach can perform multiple colonoscopy data augmentations, which improve the downstream polyp classification performance over baseline and comparison methods by up to 6%.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge