Tammy Riklin-Raviv
HyDA: Hypernetworks for Test Time Domain Adaptation in Medical Imaging Analysis
Mar 06, 2025



Abstract:Medical imaging datasets often vary due to differences in acquisition protocols, patient demographics, and imaging devices. These variations in data distribution, known as domain shift, present a significant challenge in adapting imaging analysis models for practical healthcare applications. Most current domain adaptation (DA) approaches aim either to align the distributions between the source and target domains or to learn an invariant feature space that generalizes well across all domains. However, both strategies require access to a sufficient number of examples, though not necessarily annotated, from the test domain during training. This limitation hinders the widespread deployment of models in clinical settings, where target domain data may only be accessible in real time. In this work, we introduce HyDA, a novel hypernetwork framework that leverages domain characteristics rather than suppressing them, enabling dynamic adaptation at inference time. Specifically, HyDA learns implicit domain representations and uses them to adjust model parameters on-the-fly, effectively interpolating to unseen domains. We validate HyDA on two clinically relevant applications - MRI brain age prediction and chest X-ray pathology classification - demonstrating its ability to generalize across tasks and modalities. Our code is available at TBD.
Show and Tell: Visually Explainable Deep Neural Nets via Spatially-Aware Concept Bottleneck Models
Feb 27, 2025



Abstract:Modern deep neural networks have now reached human-level performance across a variety of tasks. However, unlike humans they lack the ability to explain their decisions by showing where and telling what concepts guided them. In this work, we present a unified framework for transforming any vision neural network into a spatially and conceptually interpretable model. We introduce a spatially-aware concept bottleneck layer that projects "black-box" features of pre-trained backbone models into interpretable concept maps, without requiring human labels. By training a classification layer over this bottleneck, we obtain a self-explaining model that articulates which concepts most influenced its prediction, along with heatmaps that ground them in the input image. Accordingly, we name this method "Spatially-Aware and Label-Free Concept Bottleneck Model" (SALF-CBM). Our results show that the proposed SALF-CBM: (1) Outperforms non-spatial CBM methods, as well as the original backbone, on a variety of classification tasks; (2) Produces high-quality spatial explanations, outperforming widely used heatmap-based methods on a zero-shot segmentation task; (3) Facilitates model exploration and debugging, enabling users to query specific image regions and refine the model's decisions by locally editing its concept maps.
A Deep Ensemble Learning Approach to Lung CT Segmentation for COVID-19 Severity Assessment
Jul 05, 2022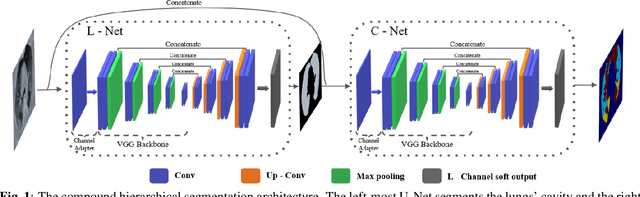
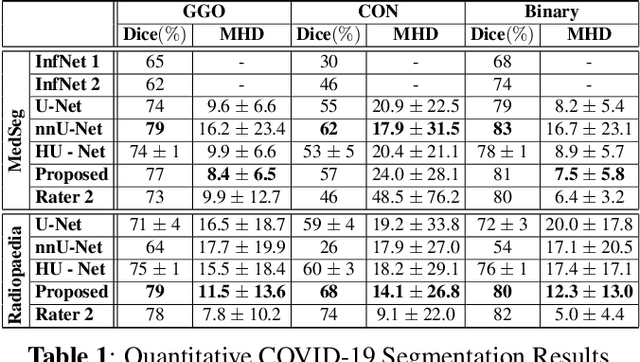
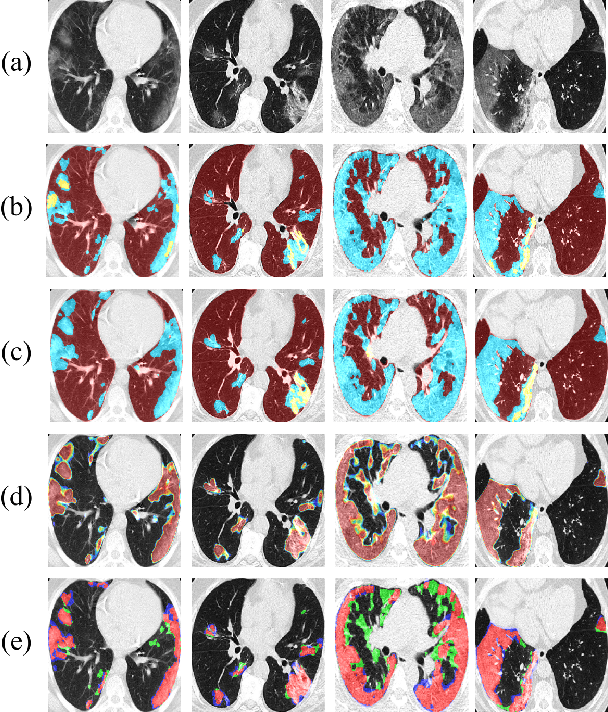
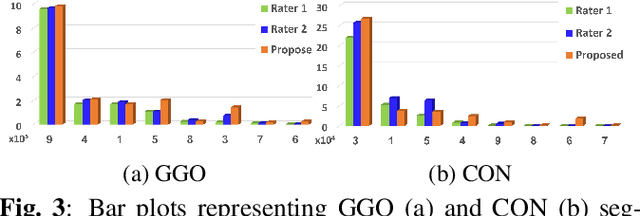
Abstract:We present a novel deep learning approach to categorical segmentation of lung CTs of COVID-19 patients. Specifically, we partition the scans into healthy lung tissues, non-lung regions, and two different, yet visually similar, pathological lung tissues, namely, ground-glass opacity and consolidation. This is accomplished via a unique, end-to-end hierarchical network architecture and ensemble learning, which contribute to the segmentation and provide a measure for segmentation uncertainty. The proposed framework achieves competitive results and outstanding generalization capabilities for three COVID-19 datasets. Our method is ranked second in a public Kaggle competition for COVID-19 CT images segmentation. Moreover, segmentation uncertainty regions are shown to correspond to the disagreements between the manual annotations of two different radiologists. Finally, preliminary promising correspondence results are shown for our private dataset when comparing the patients' COVID-19 severity scores (based on clinical measures), and the segmented lung pathologies. Code and data are available at our repository: https://github.com/talbenha/covid-seg
Graph Neural Network for Cell Tracking in Microscopy Videos
Feb 09, 2022
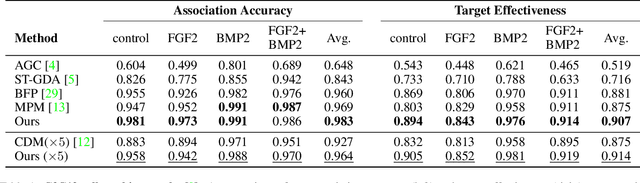

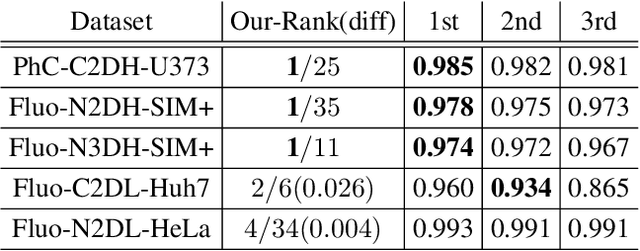
Abstract:We present a novel graph neural network (GNN) approach for cell tracking in high-throughput microscopy videos. By modeling the entire time-lapse sequence as a direct graph where cell instances are represented by its nodes and their associations by its edges, we extract the entire set of cell trajectories by looking for the maximal paths in the graph. This is accomplished by several key contributions incorporated into an end-to-end deep learning framework. We exploit a deep metric learning algorithm to extract cell feature vectors that distinguish between instances of different biological cells and assemble same cell instances. We introduce a new GNN block type which enables a mutual update of node and edge feature vectors, thus facilitating the underlying message passing process. The message passing concept, whose extent is determined by the number of GNN blocks, is of fundamental importance as it enables the `flow' of information between nodes and edges much behind their neighbors in consecutive frames. Finally, we solve an edge classification problem and use the identified active edges to construct the cells' tracks and lineage trees. We demonstrate the strengths of the proposed cell tracking approach by applying it to 2D and 3D datasets of different cell types, imaging setups, and experimental conditions. We show that our framework outperforms most of the current state-of-the-art methods.
The Role of Regularization in Shaping Weight and Node Pruning Dependency and Dynamics
Dec 07, 2020



Abstract:The pressing need to reduce the capacity of deep neural networks has stimulated the development of network dilution methods and their analysis. While the ability of $L_1$ and $L_0$ regularization to encourage sparsity is often mentioned, $L_2$ regularization is seldom discussed in this context. We present a novel framework for weight pruning by sampling from a probability function that favors the zeroing of smaller weights. In addition, we examine the contribution of $L_1$ and $L_2$ regularization to the dynamics of node pruning while optimizing for weight pruning. We then demonstrate the effectiveness of the proposed stochastic framework when used together with a weight decay regularizer on popular classification models in removing 50% of the nodes in an MLP for MNIST classification, 60% of the filters in VGG-16 for CIFAR10 classification, and on medical image models in removing 60% of the channels in a U-Net for instance segmentation and 50% of the channels in CNN model for COVID-19 detection. For these node-pruned networks, we also present competitive weight pruning results that are only slightly less accurate than the original, dense networks.
The Impact of an Inter-rater Bias on Neural Network Training
Jun 12, 2019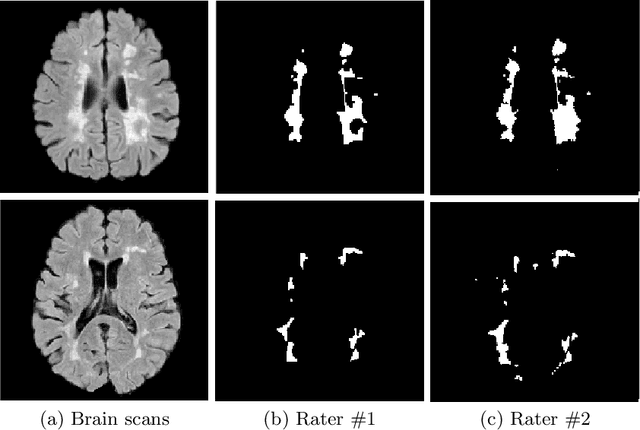



Abstract:The problem of inter-rater variability is often discussed in the context of manual labeling of medical images. It is assumed to be bypassed by automatic model-based approaches for image segmentation which are considered `objective', providing single, deterministic solutions. However, the emergence of data-driven approaches such as Deep Neural Networks (DNNs) and their application to supervised semantic segmentation - brought this issue of raters' disagreement back to the front-stage. In this paper, we highlight the issue of inter-rater bias as opposed to random inter-observer variability and demonstrate its influence on DNN training, leading to different segmentation results for the same input images. In fact, lower Dice scores are calculated if training and test segmentations are of different raters. Moreover, we demonstrate that inter-rater bias in the training examples is amplified when considering the segmentation predictions for the test data. We support our findings by showing that a classifier-DNN trained to distinguish between raters based on their manual annotations performs better when the automatic segmentation predictions rather than the raters' annotations were tested. For this study, we used the ISBI 2015 Multiple Sclerosis (MS) challenge dataset, which includes annotations by two raters with different levels of expertise. The results obtained allow us to underline a worrisome clinical implication of a DNN bias induced by an inter-rater bias during training. Specially, we show that the differences in MS-lesion load estimates increase when the volume calculations are done based on the DNNs' segmentation predictions instead of the manual annotations used for training.
DeepTract: A Probabilistic Deep Learning Framework for White Matter Fiber Tractography
Dec 12, 2018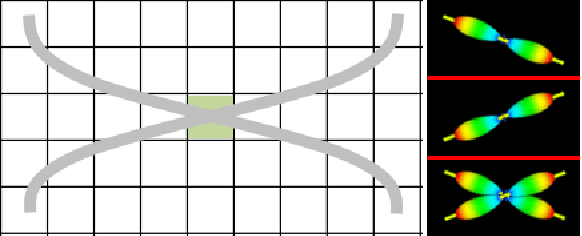
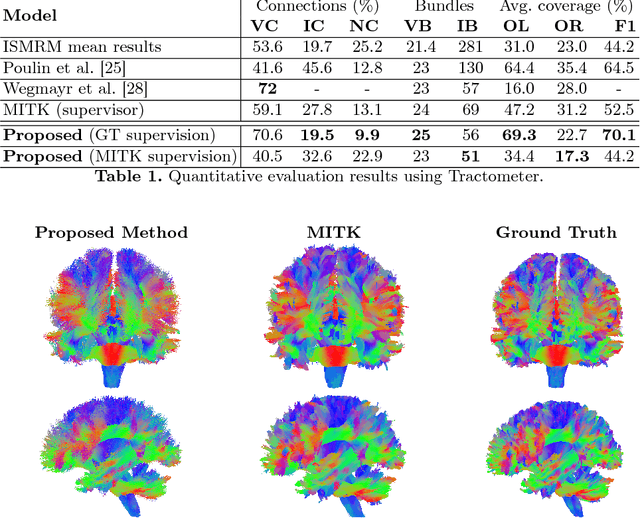


Abstract:We present DeepTract, a deep-learning framework for estimation of white matter fibers orientation and streamline tractography. We take a data-driven approach for fiber reconstruction from raw diffusion MRI, without assuming a specific diffusion model. We use a recurrent neural network for mapping sequences of diffusion-weighted imaging (DWI) values into probabilistic fiber orientation distributions. Based on these estimations, our model can perform both deterministic and probabilistic tractography on unseen DWI datasets. We quantitatively evaluate our method using the Tractometer tool, demonstrating comparable performance to state-of-the-art classical and DL-based methods. We further present qualitative results of bundle-specific probabilistic tractography of our method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge