Suhas Srinath
UnDIVE: Generalized Underwater Video Enhancement Using Generative Priors
Nov 08, 2024



Abstract:With the rise of marine exploration, underwater imaging has gained significant attention as a research topic. Underwater video enhancement has become crucial for real-time computer vision tasks in marine exploration. However, most existing methods focus on enhancing individual frames and neglect video temporal dynamics, leading to visually poor enhancements. Furthermore, the lack of ground-truth references limits the use of abundant available underwater video data in many applications. To address these issues, we propose a two-stage framework for enhancing underwater videos. The first stage uses a denoising diffusion probabilistic model to learn a generative prior from unlabeled data, capturing robust and descriptive feature representations. In the second stage, this prior is incorporated into a physics-based image formulation for spatial enhancement, while also enforcing temporal consistency between video frames. Our method enables real-time and computationally-efficient processing of high-resolution underwater videos at lower resolutions, and offers efficient enhancement in the presence of diverse water-types. Extensive experiments on four datasets show that our approach generalizes well and outperforms existing enhancement methods. Our code is available at github.com/suhas-srinath/undive.
Learning Generalizable Perceptual Representations for Data-Efficient No-Reference Image Quality Assessment
Dec 08, 2023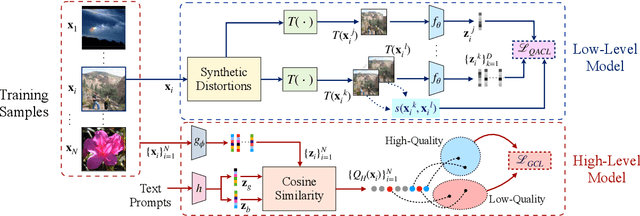
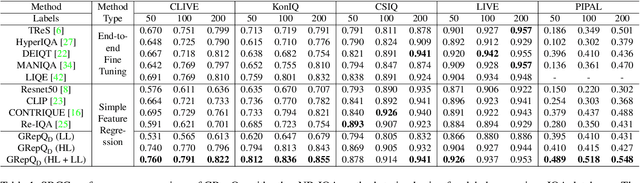
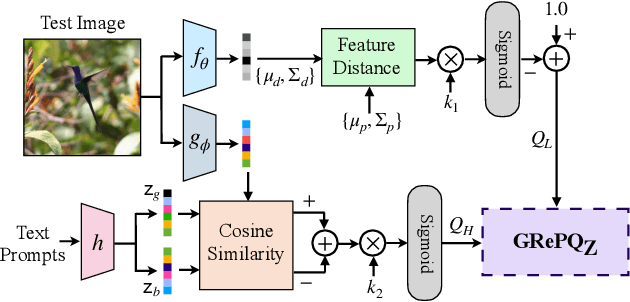
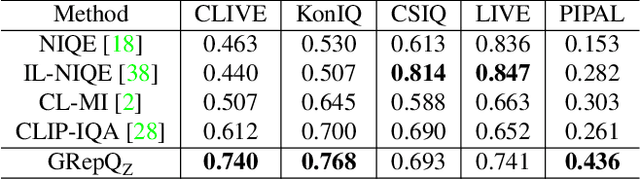
Abstract:No-reference (NR) image quality assessment (IQA) is an important tool in enhancing the user experience in diverse visual applications. A major drawback of state-of-the-art NR-IQA techniques is their reliance on a large number of human annotations to train models for a target IQA application. To mitigate this requirement, there is a need for unsupervised learning of generalizable quality representations that capture diverse distortions. We enable the learning of low-level quality features agnostic to distortion types by introducing a novel quality-aware contrastive loss. Further, we leverage the generalizability of vision-language models by fine-tuning one such model to extract high-level image quality information through relevant text prompts. The two sets of features are combined to effectively predict quality by training a simple regressor with very few samples on a target dataset. Additionally, we design zero-shot quality predictions from both pathways in a completely blind setting. Our experiments on diverse datasets encompassing various distortions show the generalizability of the features and their superior performance in the data-efficient and zero-shot settings. Code will be made available at https://github.com/suhas-srinath/GRepQ.
GenSelfDiff-HIS: Generative Self-Supervision Using Diffusion for Histopathological Image Segmentation
Sep 04, 2023Abstract:Histopathological image segmentation is a laborious and time-intensive task, often requiring analysis from experienced pathologists for accurate examinations. To reduce this burden, supervised machine-learning approaches have been adopted using large-scale annotated datasets for histopathological image analysis. However, in several scenarios, the availability of large-scale annotated data is a bottleneck while training such models. Self-supervised learning (SSL) is an alternative paradigm that provides some respite by constructing models utilizing only the unannotated data which is often abundant. The basic idea of SSL is to train a network to perform one or many pseudo or pretext tasks on unannotated data and use it subsequently as the basis for a variety of downstream tasks. It is seen that the success of SSL depends critically on the considered pretext task. While there have been many efforts in designing pretext tasks for classification problems, there haven't been many attempts on SSL for histopathological segmentation. Motivated by this, we propose an SSL approach for segmenting histopathological images via generative diffusion models in this paper. Our method is based on the observation that diffusion models effectively solve an image-to-image translation task akin to a segmentation task. Hence, we propose generative diffusion as the pretext task for histopathological image segmentation. We also propose a multi-loss function-based fine-tuning for the downstream task. We validate our method using several metrics on two publically available datasets along with a newly proposed head and neck (HN) cancer dataset containing hematoxylin and eosin (H\&E) stained images along with annotations. Codes will be made public at https://github.com/PurmaVishnuVardhanReddy/GenSelfDiff-HIS.git.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge