Sudarsun Santhiappan
PolicyBot - Reliable Question Answering over Policy Documents
Nov 17, 2025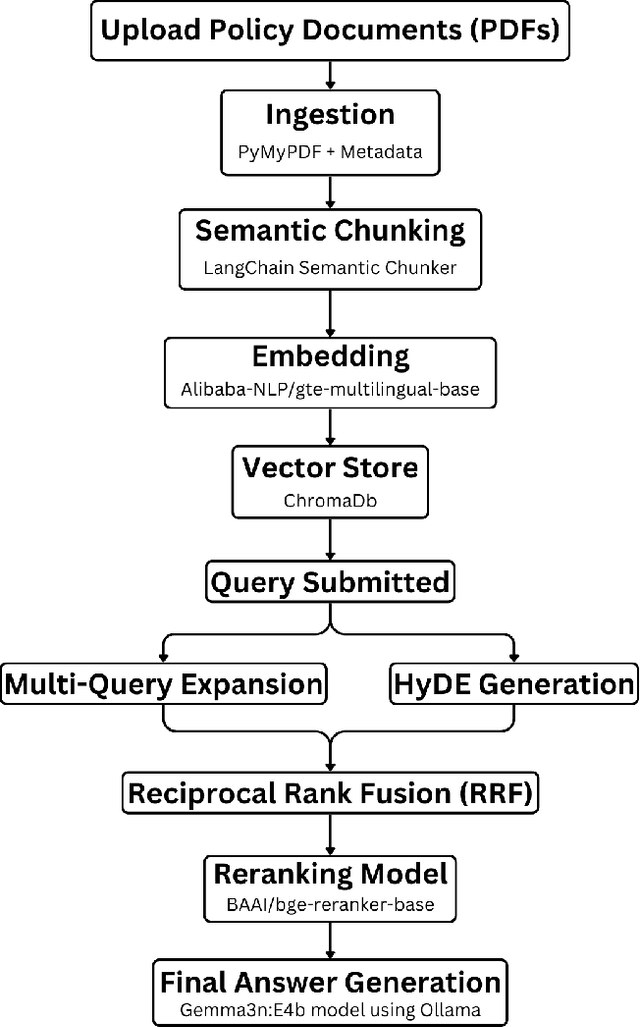
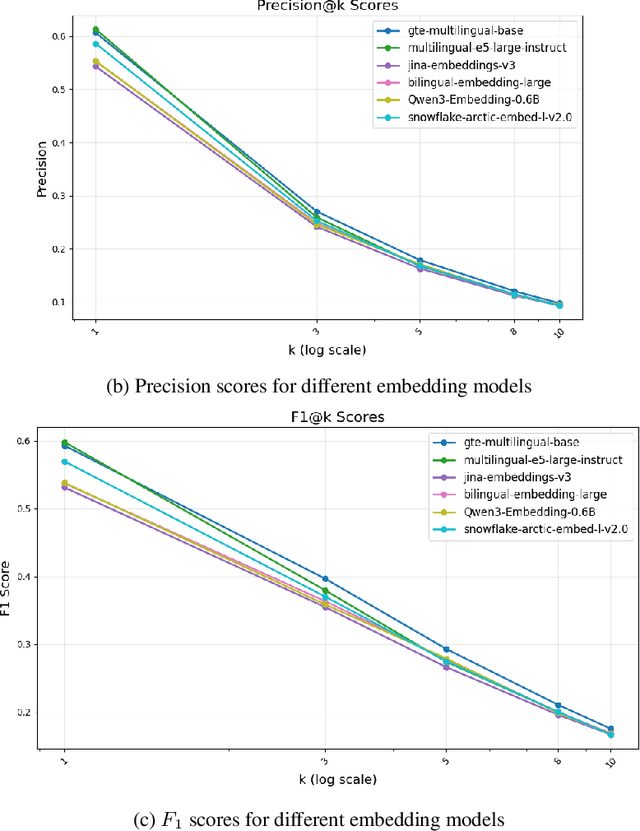
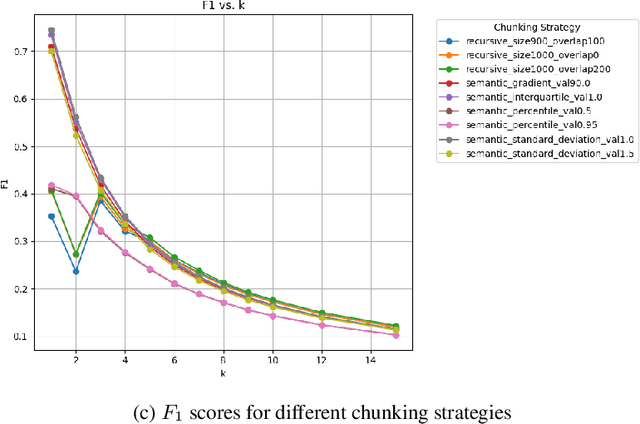
Abstract:All citizens of a country are affected by the laws and policies introduced by their government. These laws and policies serve essential functions for citizens. Such as granting them certain rights or imposing specific obligations. However, these documents are often lengthy, complex, and difficult to navigate, making it challenging for citizens to locate and understand relevant information. This work presents PolicyBot, a retrieval-augmented generation (RAG) system designed to answer user queries over policy documents with a focus on transparency and reproducibility. The system combines domain-specific semantic chunking, multilingual dense embeddings, multi-stage retrieval with reranking, and source-aware generation to provide responses grounded in the original documents. We implemented citation tracing to reduce hallucinations and improve user trust, and evaluated alternative retrieval and generation configurations to identify effective design choices. The end-to-end pipeline is built entirely with open-source tools, enabling easy adaptation to other domains requiring document-grounded question answering. This work highlights design considerations, practical challenges, and lessons learned in deploying trustworthy RAG systems for governance-related contexts.
Clustering Indices based Automatic Classification Model Selection
May 23, 2023



Abstract:Classification model selection is a process of identifying a suitable model class for a given classification task on a dataset. Traditionally, model selection is based on cross-validation, meta-learning, and user preferences, which are often time-consuming and resource-intensive. The performance of any machine learning classification task depends on the choice of the model class, the learning algorithm, and the dataset's characteristics. Our work proposes a novel method for automatic classification model selection from a set of candidate model classes by determining the empirical model-fitness for a dataset based only on its clustering indices. Clustering Indices measure the ability of a clustering algorithm to induce good quality neighborhoods with similar data characteristics. We propose a regression task for a given model class, where the clustering indices of a given dataset form the features and the dependent variable represents the expected classification performance. We compute the dataset clustering indices and directly predict the expected classification performance using the learned regressor for each candidate model class to recommend a suitable model class for dataset classification. We evaluate our model selection method through cross-validation with 60 publicly available binary class datasets and show that our top3 model recommendation is accurate for over 45 of 60 datasets. We also propose an end-to-end Automated ML system for data classification based on our model selection method. We evaluate our end-to-end system against popular commercial and noncommercial Automated ML systems using a different collection of 25 public domain binary class datasets. We show that the proposed system outperforms other methods with an excellent average rank of 1.68.
DEXTER: An end-to-end system to extract table contents from electronic medical health documents
Jul 18, 2022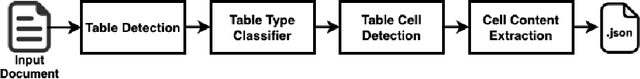


Abstract:In this paper, we propose DEXTER, an end to end system to extract information from tables present in medical health documents, such as electronic health records (EHR) and explanation of benefits (EOB). DEXTER consists of four sub-system stages: i) table detection ii) table type classification iii) cell detection; and iv) cell content extraction. We propose a two-stage transfer learning-based approach using CDeC-Net architecture along with Non-Maximal suppression for table detection. We design a conventional computer vision-based approach for table type classification and cell detection using parameterized kernels based on image size for detecting rows and columns. Finally, we extract the text from the detected cells using pre-existing OCR engine Tessaract. To evaluate our system, we manually annotated a sample of the real-world medical dataset (referred to as Meddata) consisting of wide variations of documents (in terms of appearance) covering different table structures, such as bordered, partially bordered, borderless, or coloured tables. We experimentally show that DEXTER outperforms the commercially available Amazon Textract and Microsoft Azure Form Recognizer systems on the annotated real-world medical dataset
GrabQC: Graph based Query Contextualization for automated ICD coding
Jul 14, 2022Abstract:Automated medical coding is a process of codifying clinical notes to appropriate diagnosis and procedure codes automatically from the standard taxonomies such as ICD (International Classification of Diseases) and CPT (Current Procedure Terminology). The manual coding process involves the identification of entities from the clinical notes followed by querying a commercial or non-commercial medical codes Information Retrieval (IR) system that follows the Centre for Medicare and Medicaid Services (CMS) guidelines. We propose to automate this manual process by automatically constructing a query for the IR system using the entities auto-extracted from the clinical notes. We propose \textbf{GrabQC}, a \textbf{Gra}ph \textbf{b}ased \textbf{Q}uery \textbf{C}ontextualization method that automatically extracts queries from the clinical text, contextualizes the queries using a Graph Neural Network (GNN) model and obtains the ICD Codes using an external IR system. We also propose a method for labelling the dataset for training the model. We perform experiments on two datasets of clinical text in three different setups to assert the effectiveness of our approach. The experimental results show that our proposed method is better than the compared baselines in all three settings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge