Siddharth Ramchandran
A Variational Autoencoder for Heterogeneous Temporal and Longitudinal Data
Apr 20, 2022
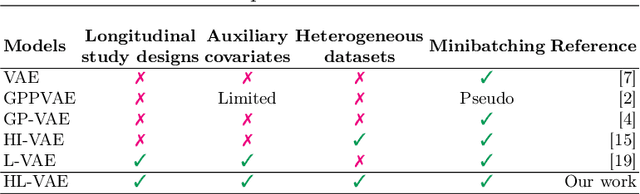

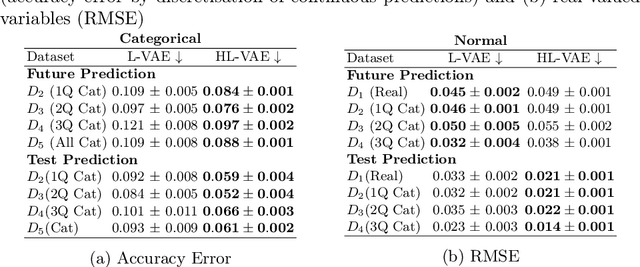
Abstract:The variational autoencoder (VAE) is a popular deep latent variable model used to analyse high-dimensional datasets by learning a low-dimensional latent representation of the data. It simultaneously learns a generative model and an inference network to perform approximate posterior inference. Recently proposed extensions to VAEs that can handle temporal and longitudinal data have applications in healthcare, behavioural modelling, and predictive maintenance. However, these extensions do not account for heterogeneous data (i.e., data comprising of continuous and discrete attributes), which is common in many real-life applications. In this work, we propose the heterogeneous longitudinal VAE (HL-VAE) that extends the existing temporal and longitudinal VAEs to heterogeneous data. HL-VAE provides efficient inference for high-dimensional datasets and includes likelihood models for continuous, count, categorical, and ordinal data while accounting for missing observations. We demonstrate our model's efficacy through simulated as well as clinical datasets, and show that our proposed model achieves competitive performance in missing value imputation and predictive accuracy.
Learning Conditional Variational Autoencoders with Missing Covariates
Mar 02, 2022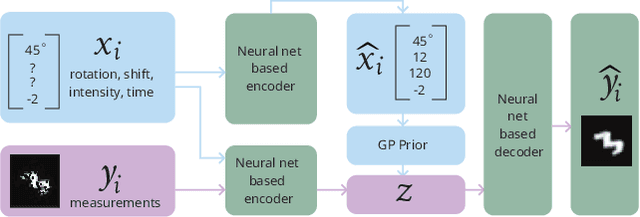



Abstract:Conditional variational autoencoders (CVAEs) are versatile deep generative models that extend the standard VAE framework by conditioning the generative model with auxiliary covariates. The original CVAE model assumes that the data samples are independent, whereas more recent conditional VAE models, such as the Gaussian process (GP) prior VAEs, can account for complex correlation structures across all data samples. While several methods have been proposed to learn standard VAEs from partially observed datasets, these methods fall short for conditional VAEs. In this work, we propose a method to learn conditional VAEs from datasets in which auxiliary covariates can contain missing values as well. The proposed method augments the conditional VAEs with a prior distribution for the missing covariates and estimates their posterior using amortised variational inference. At training time, our method marginalises the uncertainty associated with the missing covariates while simultaneously maximising the evidence lower bound. We develop computationally efficient methods to learn CVAEs and GP prior VAEs that are compatible with mini-batching. Our experiments on simulated datasets as well as on a clinical trial study show that the proposed method outperforms previous methods in learning conditional VAEs from non-temporal, temporal, and longitudinal datasets.
Longitudinal Variational Autoencoder
Jun 17, 2020

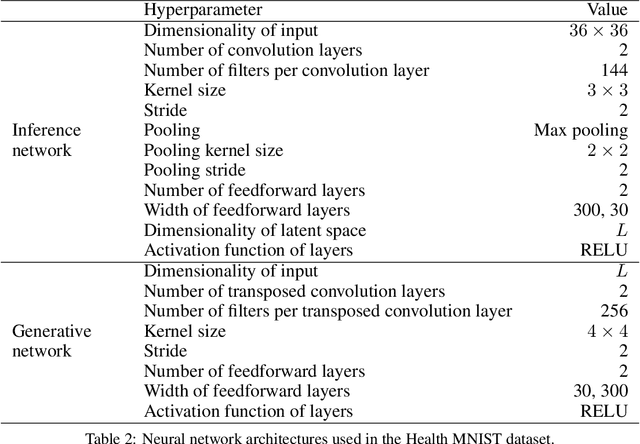
Abstract:Longitudinal datasets measured repeatedly over time from individual subjects, arise in many biomedical, psychological, social, and other studies. Such multivariate time-series are often high-dimensional and contain missing values. A common approach to analyse this kind of data is to learn a low-dimensional representation using variational autoencoders (VAEs). However, standard VAEs assume that the learned representations are i.i.d., and fail to capture the correlations between the data samples. We propose a novel deep generative model, Longitudinal VAE (L-VAE), that uses a multi-output additive Gaussian process (GP) prior to extend the VAE's capability to learn structured low-dimensional representations imposed by auxiliary covariate information, and also derive a new divergence upper bound for such GPs. Our approach can simultaneously accommodate both time-varying shared and random effects, produce structured low-dimensional representations, disentangle effects of individual covariates or their interactions, and achieve highly accurate predictive performance. We compare our model against previous methods on synthetic and clinical datasets, and demonstrate the state-of-the-art performance in data imputation, reconstruction, and long-term prediction tasks.
Latent Gaussian process with composite likelihoods for data-driven disease stratification
Sep 04, 2019
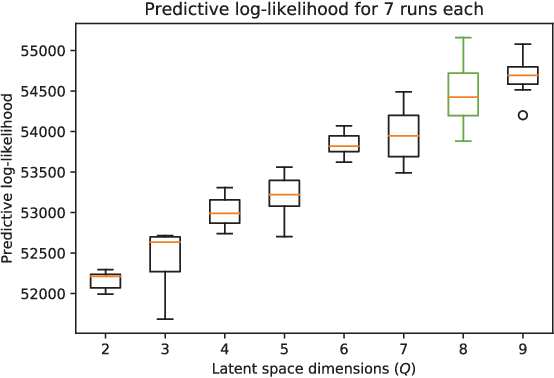
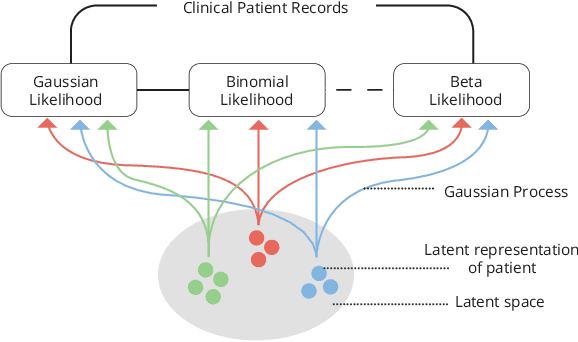
Abstract:Data-driven techniques for identifying disease subtypes using medical records can greatly benefit the management of patients' health and unravel the underpinnings of diseases. Clinical patient records are typically collected from disparate sources and result in high-dimensional data comprising of multiple likelihoods with noisy and missing values. Probabilistic methods capable of analysing large-scale patient records have a central role in biomedical research and are expected to become even more important when data-driven personalised medicine will be established in clinical practise. In this work we propose an unsupervised, generative model that can identify clustering among patients in a latent space while making use of all available data (i.e. in a heterogeneous data setting with noisy and missing values). We make use of the Gaussian process latent variable models (GPLVM) and deep neural networks to create a non-linear dimensionality reduction technique for heterogeneous data. The effectiveness of our model is demonstrated on clinical data of Parkinson's disease patients treated at the HUS Helsinki University Hospital. We demonstrate sub-groups from the heterogeneous patient data, evaluate the robustness of the findings, and interpret cluster characteristics.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge