Latent Gaussian process with composite likelihoods for data-driven disease stratification
Paper and Code
Sep 04, 2019
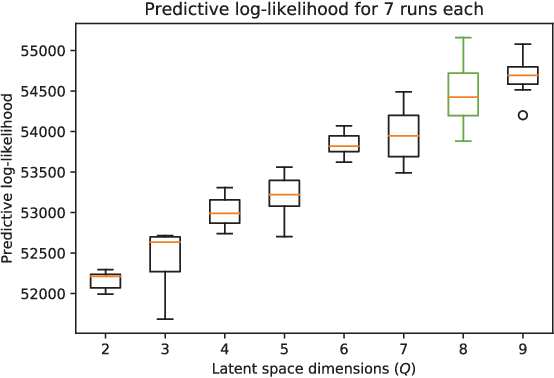
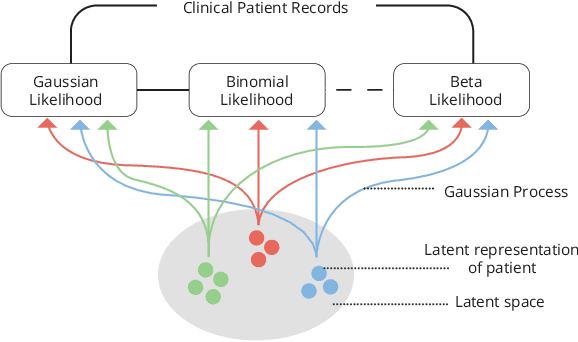
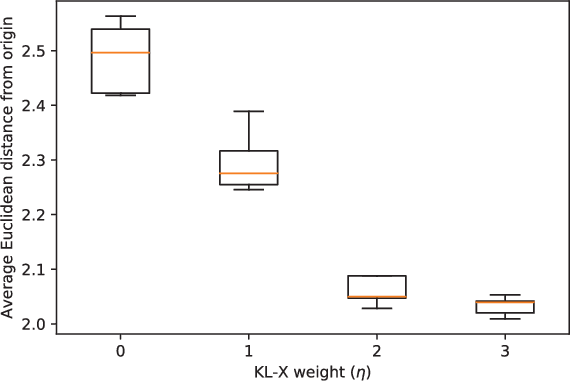
Data-driven techniques for identifying disease subtypes using medical records can greatly benefit the management of patients' health and unravel the underpinnings of diseases. Clinical patient records are typically collected from disparate sources and result in high-dimensional data comprising of multiple likelihoods with noisy and missing values. Probabilistic methods capable of analysing large-scale patient records have a central role in biomedical research and are expected to become even more important when data-driven personalised medicine will be established in clinical practise. In this work we propose an unsupervised, generative model that can identify clustering among patients in a latent space while making use of all available data (i.e. in a heterogeneous data setting with noisy and missing values). We make use of the Gaussian process latent variable models (GPLVM) and deep neural networks to create a non-linear dimensionality reduction technique for heterogeneous data. The effectiveness of our model is demonstrated on clinical data of Parkinson's disease patients treated at the HUS Helsinki University Hospital. We demonstrate sub-groups from the heterogeneous patient data, evaluate the robustness of the findings, and interpret cluster characteristics.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge