Shuyue Guan
CaraNet: Context Axial Reverse Attention Network for Segmentation of Small Medical Objects
Jan 31, 2023Abstract:Segmenting medical images accurately and reliably is important for disease diagnosis and treatment. It is a challenging task because of the wide variety of objects' sizes, shapes, and scanning modalities. Recently, many convolutional neural networks (CNN) have been designed for segmentation tasks and achieved great success. Few studies, however, have fully considered the sizes of objects, and thus most demonstrate poor performance for small objects segmentation. This can have a significant impact on the early detection of diseases. This paper proposes a Context Axial Reverse Attention Network (CaraNet) to improve the segmentation performance on small objects compared with several recent state-of-the-art models. CaraNet applies axial reserve attention (ARA) and channel-wise feature pyramid (CFP) module to dig feature information of small medical object. And we evaluate our model by six different measurement metrics. We test our CaraNet on brain tumor (BraTS 2018) and polyp (Kvasir-SEG, CVC-ColonDB, CVC-ClinicDB, CVC-300, and ETIS-LaribPolypDB) segmentation datasets. Our CaraNet achieves the top-rank mean Dice segmentation accuracy, and results show a distinct advantage of CaraNet in the segmentation of small medical objects.
Informing selection of performance metrics for medical image segmentation evaluation using configurable synthetic errors
Dec 30, 2022



Abstract:Machine learning-based segmentation in medical imaging is widely used in clinical applications from diagnostics to radiotherapy treatment planning. Segmented medical images with ground truth are useful for investigating the properties of different segmentation performance metrics to inform metric selection. Regular geometrical shapes are often used to synthesize segmentation errors and illustrate properties of performance metrics, but they lack the complexity of anatomical variations in real images. In this study, we present a tool to emulate segmentations by adjusting the reference (truth) masks of anatomical objects extracted from real medical images. Our tool is designed to modify the defined truth contours and emulate different types of segmentation errors with a set of user-configurable parameters. We defined the ground truth objects from 230 patient images in the Glioma Image Segmentation for Radiotherapy (GLIS-RT) database. For each object, we used our segmentation synthesis tool to synthesize 10 versions of segmentation (i.e., 10 simulated segmentors or algorithms), where each version has a pre-defined combination of segmentation errors. We then applied 20 performance metrics to evaluate all synthetic segmentations. We demonstrated the properties of these metrics, including their ability to capture specific types of segmentation errors. By analyzing the intrinsic properties of these metrics and categorizing the segmentation errors, we are working toward the goal of developing a decision-tree tool for assisting in the selection of segmentation performance metrics.
A Sneak Attack on Segmentation of Medical Images Using Deep Neural Network Classifiers
Jan 28, 2022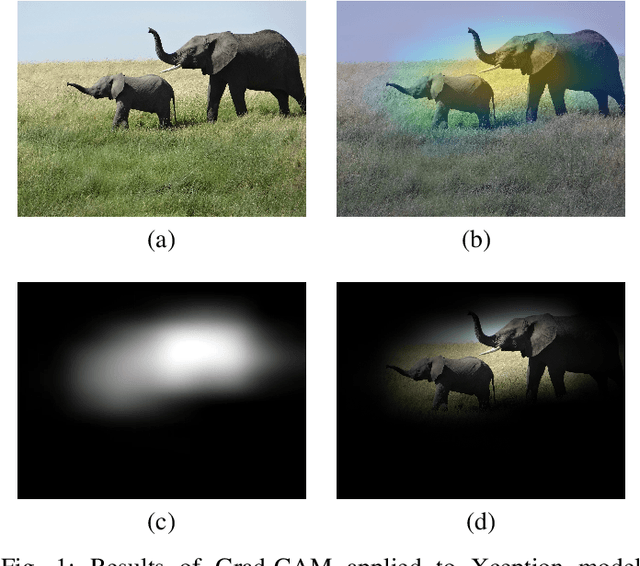


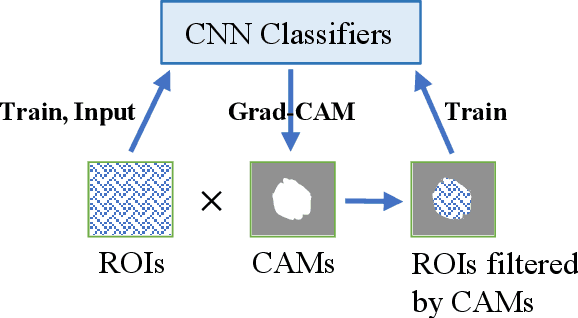
Abstract:Instead of using current deep-learning segmentation models (like the UNet and variants), we approach the segmentation problem using trained Convolutional Neural Network (CNN) classifiers, which automatically extract important features from images for classification. Those extracted features can be visualized and formed into heatmaps using Gradient-weighted Class Activation Mapping (Grad-CAM). This study tested whether the heatmaps could be used to segment the classified targets. We also proposed an evaluation method for the heatmaps; that is, to re-train the CNN classifier using images filtered by heatmaps and examine its performance. We used the mean-Dice coefficient to evaluate segmentation results. Results from our experiments show that heatmaps can locate and segment partial tumor areas. But use of only the heatmaps from CNN classifiers may not be an optimal approach for segmentation. We have verified that the predictions of CNN classifiers mainly depend on tumor areas, and dark regions in Grad-CAM's heatmaps also contribute to classification.
A Novel Intrinsic Measure of Data Separability
Sep 11, 2021
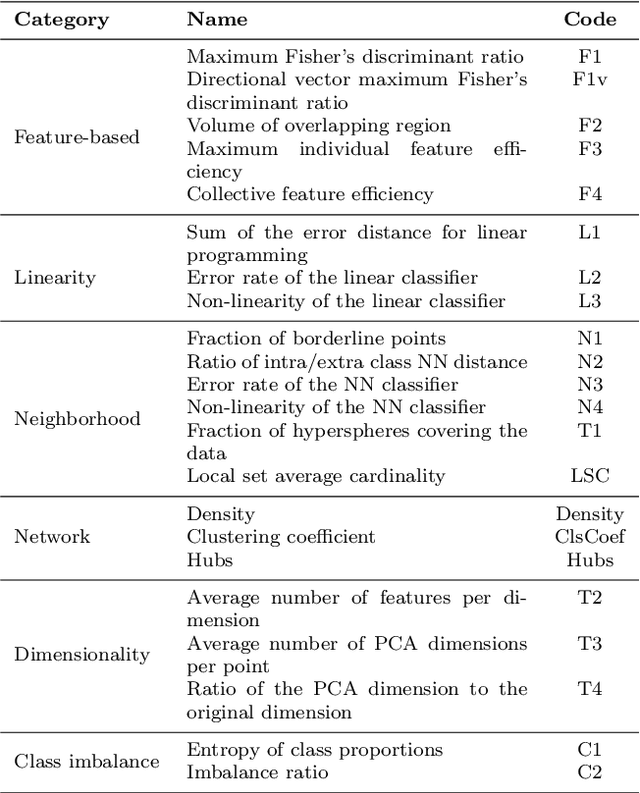

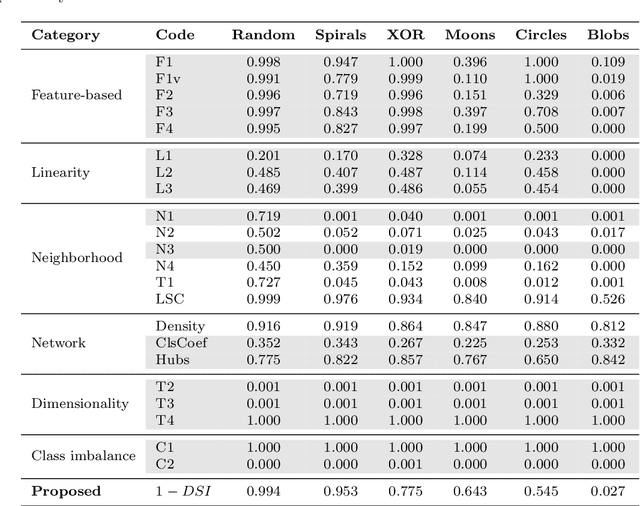
Abstract:In machine learning, the performance of a classifier depends on both the classifier model and the separability/complexity of datasets. To quantitatively measure the separability of datasets, we create an intrinsic measure -- the Distance-based Separability Index (DSI), which is independent of the classifier model. We consider the situation in which different classes of data are mixed in the same distribution to be the most difficult for classifiers to separate. We then formally show that the DSI can indicate whether the distributions of datasets are identical for any dimensionality. And we verify the DSI to be an effective separability measure by comparing to several state-of-the-art separability/complexity measures using synthetic and real datasets. Having demonstrated the DSI's ability to compare distributions of samples, we also discuss some of its other promising applications, such as measuring the performance of generative adversarial networks (GANs) and evaluating the results of clustering methods.
A Distance-based Separability Measure for Internal Cluster Validation
Jun 17, 2021
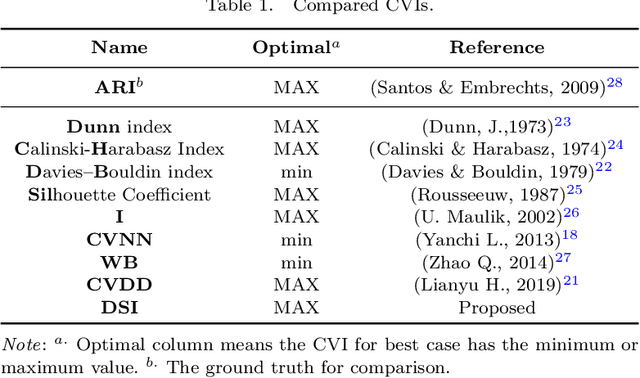
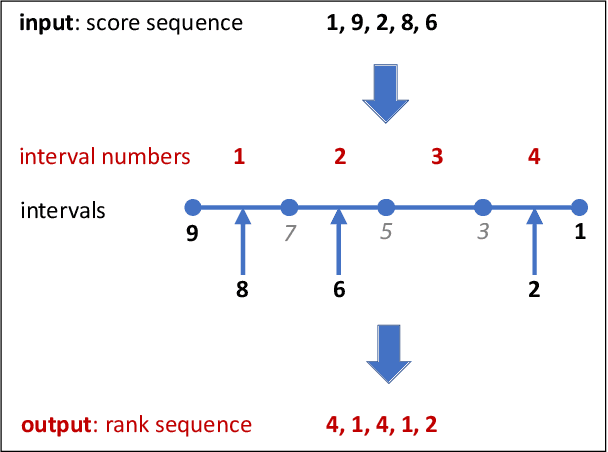
Abstract:To evaluate clustering results is a significant part of cluster analysis. Since there are no true class labels for clustering in typical unsupervised learning, many internal cluster validity indices (CVIs), which use predicted labels and data, have been created. Without true labels, to design an effective CVI is as difficult as to create a clustering method. And it is crucial to have more CVIs because there are no universal CVIs that can be used to measure all datasets and no specific methods of selecting a proper CVI for clusters without true labels. Therefore, to apply a variety of CVIs to evaluate clustering results is necessary. In this paper, we propose a novel internal CVI -- the Distance-based Separability Index (DSI), based on a data separability measure. We compared the DSI with eight internal CVIs including studies from early Dunn (1974) to most recent CVDD (2019) and an external CVI as ground truth, by using clustering results of five clustering algorithms on 12 real and 97 synthetic datasets. Results show DSI is an effective, unique, and competitive CVI to other compared CVIs. We also summarized the general process to evaluate CVIs and created the rank-difference metric for comparison of CVIs' results.
CFPNet-M: A Light-Weight Encoder-Decoder Based Network for Multimodal Biomedical Image Real-Time Segmentation
May 30, 2021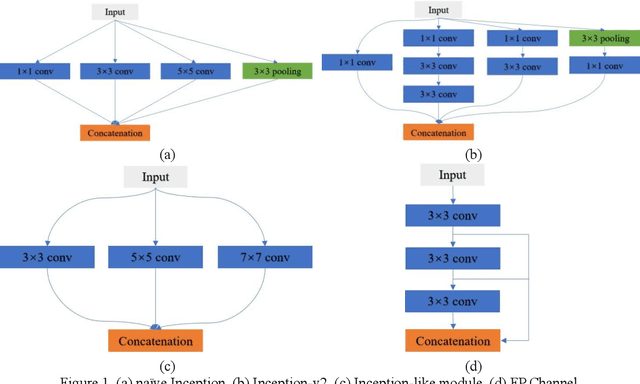
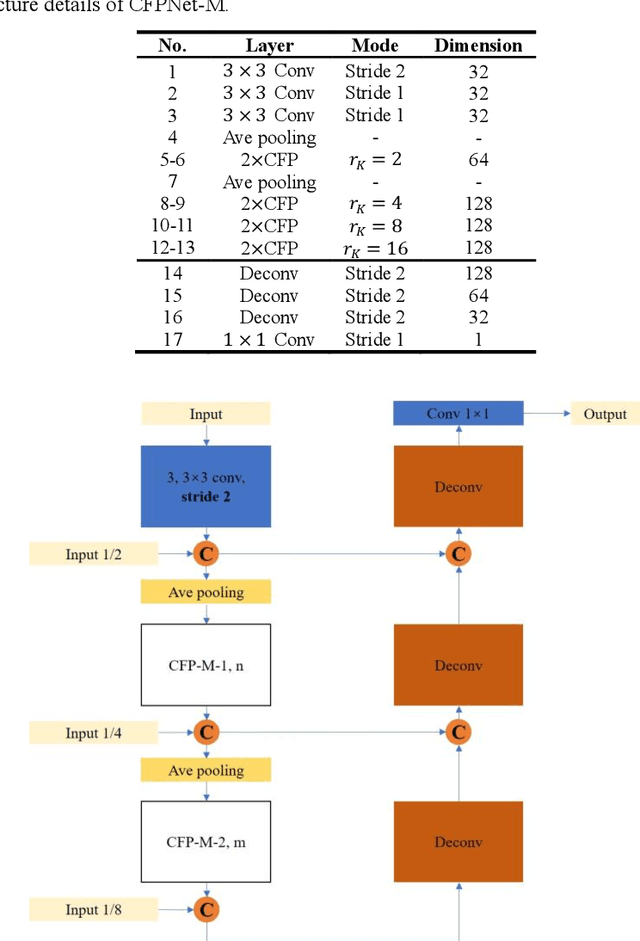
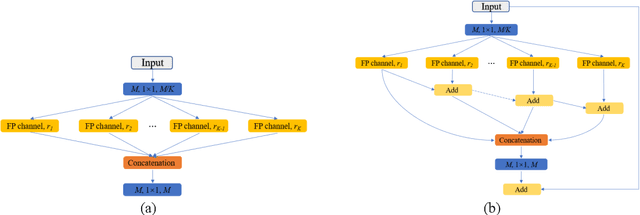

Abstract:Currently, developments of deep learning techniques are providing instrumental to identify, classify, and quantify patterns in medical images. Segmentation is one of the important applications in medical image analysis. In this regard, U-Net is the predominant approach to medical image segmentation tasks. However, we found that those U-Net based models have limitations in several aspects, for example, millions of parameters in the U-Net consuming considerable computation resource and memory, lack of global information, and missing some tough objects. Therefore, we applied two modifications to improve the U-Net model: 1) designed and added the dilated channel-wise CNN module, 2) simplified the U shape network. Based on these two modifications, we proposed a novel light-weight architecture -- Channel-wise Feature Pyramid Network for Medicine (CFPNet-M). To evaluate our method, we selected five datasets with different modalities: thermography, electron microscopy, endoscopy, dermoscopy, and digital retinal images. And we compared its performance with several models having different parameter scales. This paper also involves our previous studies of DC-UNet and some commonly used light-weight neural networks. We applied the Tanimoto similarity instead of the Jaccard index for gray-level image measurements. By comparison, CFPNet-M achieves comparable segmentation results on all five medical datasets with only 0.65 million parameters, which is about 2% of U-Net, and 8.8 MB memory. Meanwhile, the inference speed can reach 80 FPS on a single RTX 2070Ti GPU with the 256 by 192 pixels input size.
Understanding the Ability of Deep Neural Networks to Count Connected Components in Images
Jan 05, 2021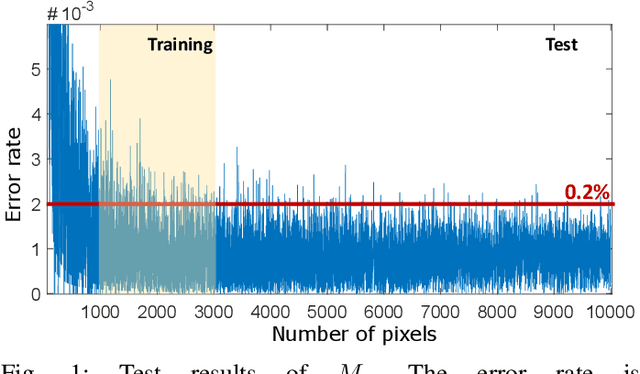


Abstract:Humans can count very fast by subitizing, but slow substantially as the number of objects increases. Previous studies have shown a trained deep neural network (DNN) detector can count the number of objects in an amount of time that increases slowly with the number of objects. Such a phenomenon suggests the subitizing ability of DNNs, and unlike humans, it works equally well for large numbers. Many existing studies have successfully applied DNNs to object counting, but few studies have studied the subitizing ability of DNNs and its interpretation. In this paper, we found DNNs do not have the ability to generally count connected components. We provided experiments to support our conclusions and explanations to understand the results and phenomena of these experiments. We proposed three ML-learnable characteristics to verify learnable problems for ML models, such as DNNs, and explain why DNNs work for specific counting problems but cannot generally count connected components.
Segmentation of Infrared Breast Images Using MultiResUnet Neural Network
Oct 31, 2020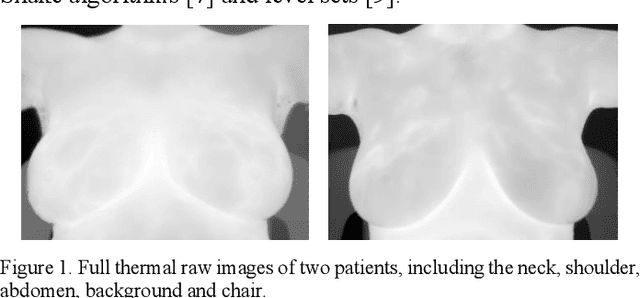


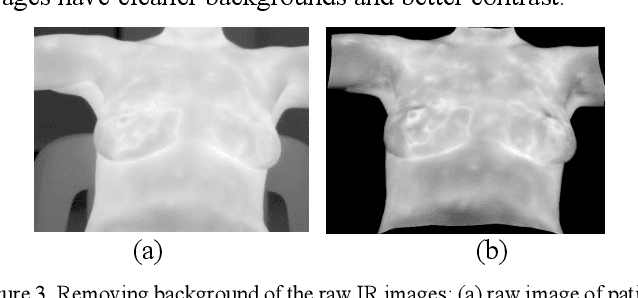
Abstract:Breast cancer is the second leading cause of death for women in the U.S. Early detection of breast cancer is key to higher survival rates of breast cancer patients. We are investigating infrared (IR) thermography as a noninvasive adjunct to mammography for breast cancer screening. IR imaging is radiation-free, pain-free, and non-contact. Automatic segmentation of the breast area from the acquired full-size breast IR images will help limit the area for tumor search, as well as reduce the time and effort costs of manual segmentation. Autoencoder-like convolutional and deconvolutional neural networks (C-DCNN) had been applied to automatically segment the breast area in IR images in previous studies. In this study, we applied a state-of-the-art deep-learning segmentation model, MultiResUnet, which consists of an encoder part to capture features and a decoder part for precise localization. It was used to segment the breast area by using a set of breast IR images, collected in our pilot study by imaging breast cancer patients and normal volunteers with a thermal infrared camera (N2 Imager). The database we used has 450 images, acquired from 14 patients and 16 volunteers. We used a thresholding method to remove interference in the raw images and remapped them from the original 16-bit to 8-bit, and then cropped and segmented the 8-bit images manually. Experiments using leave-one-out cross-validation (LOOCV) and comparison with the ground-truth images by using Tanimoto similarity show that the average accuracy of MultiResUnet is 91.47%, which is about 2% higher than that of the autoencoder. MultiResUnet offers a better approach to segment breast IR images than our previous model.
The estimation of training accuracy for two-layer neural networks on random datasets without training
Oct 26, 2020
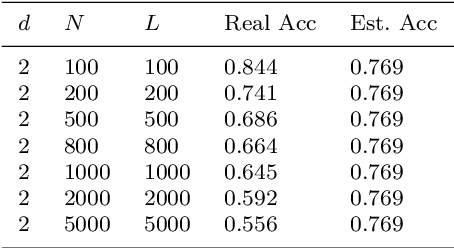

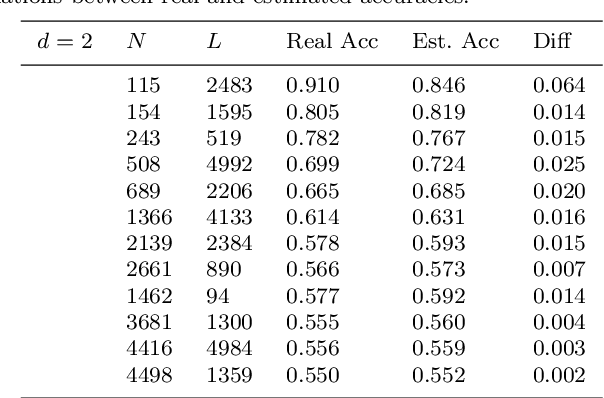
Abstract:Although the neural network (NN) technique plays an important role in machine learning, understanding the mechanism of NN models and the transparency of deep learning still require more basic research. In this study we propose a novel theory based on space partitioning to estimate the approximate training accuracy for two-layer neural networks on random datasets without training. There appear to be no other studies that have proposed a method to estimate training accuracy without using input data or trained models. Our method estimates the training accuracy for two-layer fully-connected neural networks on two-class random datasets using only three arguments: the dimensionality of inputs (d), the number of inputs (N), and the number of neurons in the hidden layer (L). We have verified our method using real training accuracies in our experiments. The results indicate that the method will work for any dimension, and the proposed theory could extend also to estimate deeper NN models. This study may provide a starting point for a new way for researchers to make progress on the difficult problem of understanding deep learning.
Analysis of Generalizability of Deep Neural Networks Based on the Complexity of Decision Boundary
Sep 16, 2020



Abstract:For supervised learning models, the analysis of generalization ability (generalizability) is vital because the generalizability expresses how well a model will perform on unseen data. Traditional generalization methods, such as the VC dimension, do not apply to deep neural network (DNN) models. Thus, new theories to explain the generalizability of DNNs are required. In this study, we hypothesize that the DNN with a simpler decision boundary has better generalizability by the law of parsimony (Occam's Razor). We create the decision boundary complexity (DBC) score to define and measure the complexity of decision boundary of DNNs. The idea of the DBC score is to generate data points (called adversarial examples) on or near the decision boundary. Our new approach then measures the complexity of the boundary using the entropy of eigenvalues of these data. The method works equally well for high-dimensional data. We use training data and the trained model to compute the DBC score. And, the ground truth for model's generalizability is its test accuracy. Experiments based on the DBC score have verified our hypothesis. The DBC is shown to provide an effective method to measure the complexity of a decision boundary and gives a quantitative measure of the generalizability of DNNs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge