Shubo Wang
Breast Cancer Induced Bone Osteolysis Prediction Using Temporal Variational Auto-Encoders
Mar 28, 2022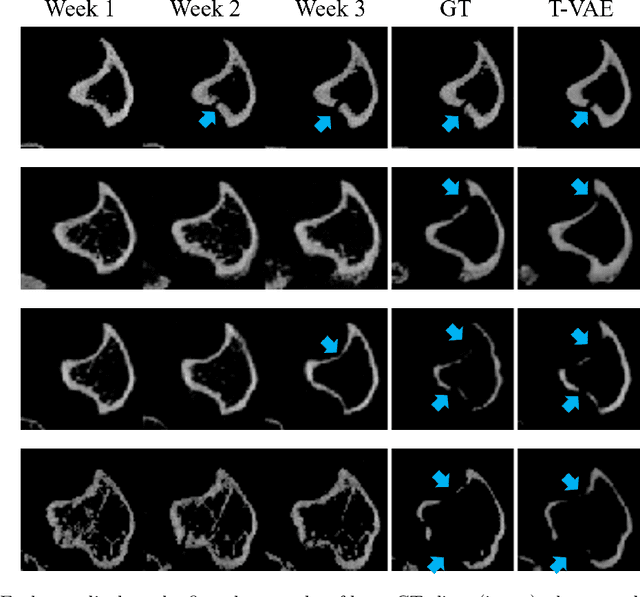
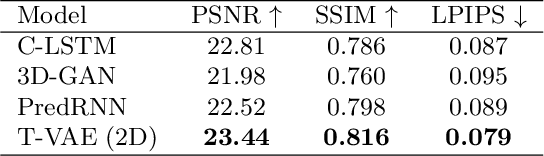
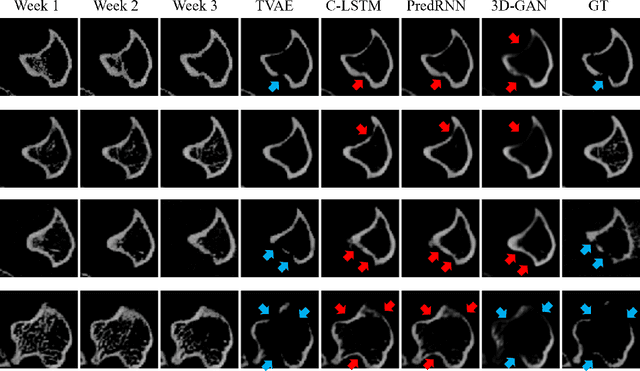

Abstract:Objective and Impact Statement. We adopt a deep learning model for bone osteolysis prediction on computed tomography (CT) images of murine breast cancer bone metastases. Given the bone CT scans at previous time steps, the model incorporates the bone-cancer interactions learned from the sequential images and generates future CT images. Its ability of predicting the development of bone lesions in cancer-invading bones can assist in assessing the risk of impending fractures and choosing proper treatments in breast cancer bone metastasis. Introduction. Breast cancer often metastasizes to bone, causes osteolytic lesions, and results in skeletal related events (SREs) including severe pain and even fatal fractures. Although current imaging techniques can detect macroscopic bone lesions, predicting the occurrence and progression of bone lesions remains a challenge. Methods. We adopt a temporal variational auto-encoder (T-VAE) model that utilizes a combination of variational auto-encoders and long short-term memory networks to predict bone lesion emergence on our micro-CT dataset containing sequential images of murine tibiae. Given the CT scans of murine tibiae at early weeks, our model can learn the distribution of their future states from data. Results. We test our model against other deep learning-based prediction models on the bone lesion progression prediction task. Our model produces much more accurate predictions than existing models under various evaluation metrics. Conclusion. We develop a deep learning framework that can accurately predict and visualize the progression of osteolytic bone lesions. It will assist in planning and evaluating treatment strategies to prevent SREs in breast cancer patients.
A Prior Knowledge Based Tumor and Tumoral Subregion Segmentation Tool for Pediatric Brain Tumors
Sep 30, 2021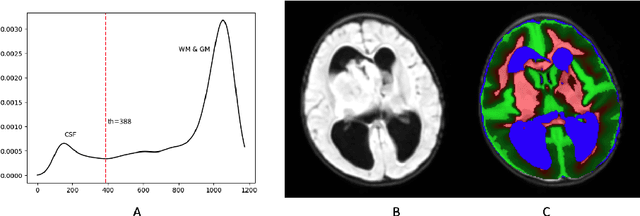
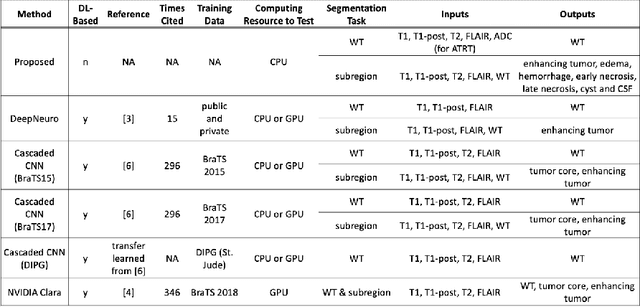
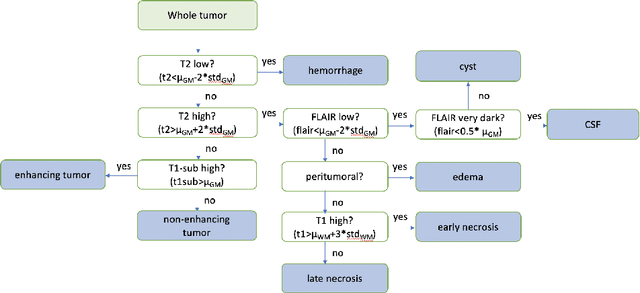
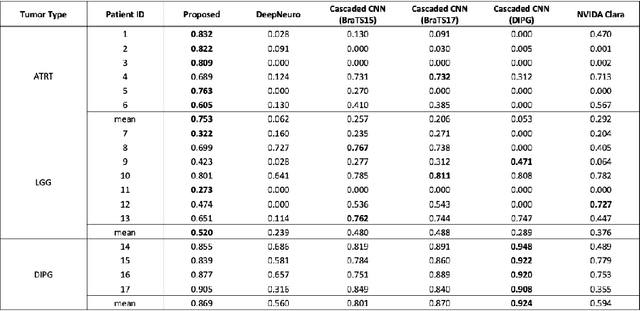
Abstract:In the past few years, deep learning (DL) models have drawn great attention and shown superior performance on brain tumor and subregion segmentation tasks. However, the success is limited to segmentation of adult gliomas, where sufficient data have been collected, manually labeled, and published for training DL models. It is still challenging to segment pediatric tumors, because the appearances are different from adult gliomas. Hence, directly applying a pretained DL model on pediatric data usually generates unacceptable results. Because pediatric data is very limited, both labeled and unlabeled, we present a brain tumor segmentation model that is based on knowledge rather than learning from data. We also provide segmentation of more subregions for super heterogeneous tumor like atypical teratoid rhabdoid tumor (ATRT). Our proposed approach showed superior performance on both whole tumor and subregion segmentation tasks to DL based models on our pediatric data when training data is not available for transfer learning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge