Silu Zhang
Automatic Detection and Segmentation of Postoperative Cerebellar Damage Based on Normalization
Mar 03, 2022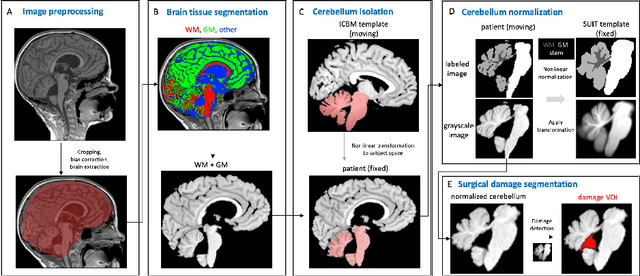
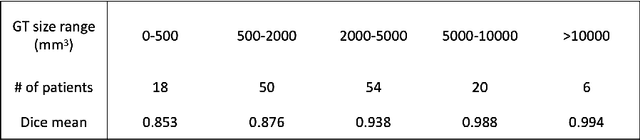
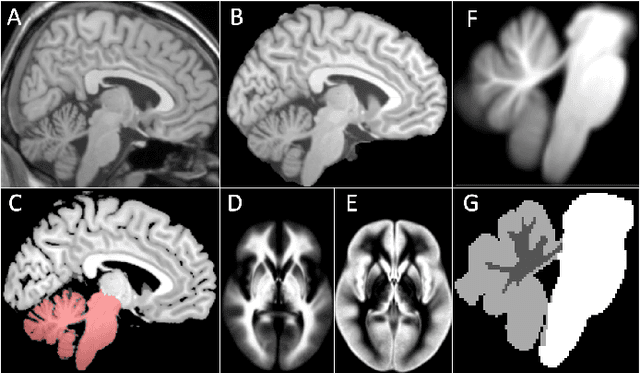

Abstract:Surgical resection is a common procedure in the treatment of pediatric posterior fossa tumors. However, surgical damage is often unavoidable and its association with postoperative complications is not well understood. A reliable localization and measure of cerebellar damage is fundamental to study the relationship between the damaged cerebellar regions and postoperative neurological outcomes. Existing cerebellum normalization methods are not reliable on postoperative scans, therefore current approaches to measure surgical damage rely on manual labelling. In this work, we develop a robust algorithm to automatically detect and measure cerebellum damage due to surgery using postoperative 3D T1 magnetic resonance imaging. In our proposed approach, normal brain tissues are first segmented using a Bayesian algorithm customized for postoperative scans. Next, the cerebellum is isolated by nonlinear registration of a whole brain template to the native space. The isolated cerebellum is then normalized into the spatially unbiased atlas (SUIT) space using anatomical information derived from the previous step. Finally, the damage is detected in the atlas space by comparing the normalized cerebellum and the SUIT template. We evaluated our damage detection tool on postoperative scans of 153 patients diagnosed with medulloblastoma based on inspection by human expects. We also designed a simulation to test the proposed approach without human intervention. Our results show that the proposed approach has superior performance on various scenarios.
A Prior Knowledge Based Tumor and Tumoral Subregion Segmentation Tool for Pediatric Brain Tumors
Sep 30, 2021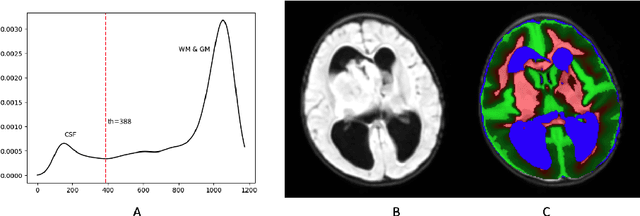
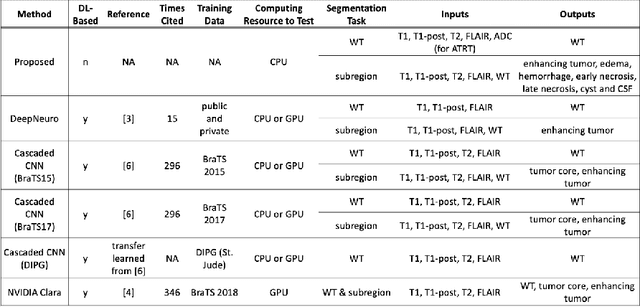
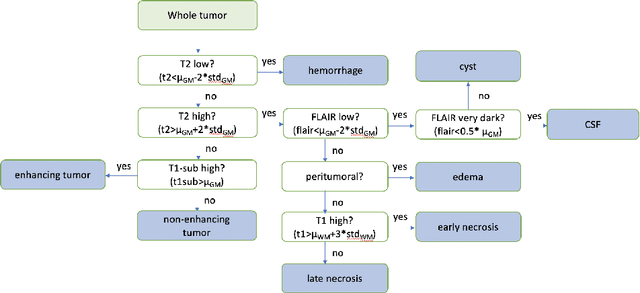
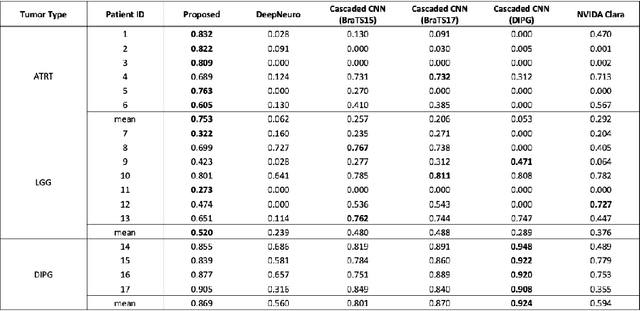
Abstract:In the past few years, deep learning (DL) models have drawn great attention and shown superior performance on brain tumor and subregion segmentation tasks. However, the success is limited to segmentation of adult gliomas, where sufficient data have been collected, manually labeled, and published for training DL models. It is still challenging to segment pediatric tumors, because the appearances are different from adult gliomas. Hence, directly applying a pretained DL model on pediatric data usually generates unacceptable results. Because pediatric data is very limited, both labeled and unlabeled, we present a brain tumor segmentation model that is based on knowledge rather than learning from data. We also provide segmentation of more subregions for super heterogeneous tumor like atypical teratoid rhabdoid tumor (ATRT). Our proposed approach showed superior performance on both whole tumor and subregion segmentation tasks to DL based models on our pediatric data when training data is not available for transfer learning.
Estimating Feature-Label Dependence Using Gini Distance Statistics
Jun 05, 2019



Abstract:Identifying statistical dependence between the features and the label is a fundamental problem in supervised learning. This paper presents a framework for estimating dependence between numerical features and a categorical label using generalized Gini distance, an energy distance in reproducing kernel Hilbert spaces (RKHS). Two Gini distance based dependence measures are explored: Gini distance covariance and Gini distance correlation. Unlike Pearson covariance and correlation, which do not characterize independence, the above Gini distance based measures define dependence as well as independence of random variables. The test statistics are simple to calculate and do not require probability density estimation. Uniform convergence bounds and asymptotic bounds are derived for the test statistics. Comparisons with distance covariance statistics are provided. It is shown that Gini distance statistics converge faster than distance covariance statistics in the uniform convergence bounds, hence tighter upper bounds on both Type I and Type II errors. Moreover, the probability of Gini distance covariance statistic under-performing the distance covariance statistic in Type II error decreases to 0 exponentially with the increase of the sample size. Extensive experimental results are presented to demonstrate the performance of the proposed method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge