Shreyas Sharma
EvolveMT: an Ensemble MT Engine Improving Itself with Usage Only
Jun 20, 2023Abstract:This paper presents EvolveMT for efficiently combining multiple machine translation (MT) engines. The proposed system selects the output from a single engine for each segment by utilizing online learning techniques to predict the most suitable system for every translation request. A neural quality estimation metric supervises the method without requiring reference translations. The online learning capability of this system allows for dynamic adaptation to alterations in the domain or machine translation engines, thereby obviating the necessity for additional training. EvolveMT selects a subset of translation engines to be called based on the source sentence features. The degree of exploration is configurable according to the desired quality-cost trade-off. Results from custom datasets demonstrate that EvolveMT achieves similar translation accuracy at a lower cost than selecting the best translation of each segment from all translations using an MT quality estimator. To our knowledge, EvolveMT is the first meta MT system that adapts itself after deployment to incoming translation requests from the production environment without needing costly retraining on human feedback.
Efficient Machine Translation Corpus Generation
Jun 20, 2023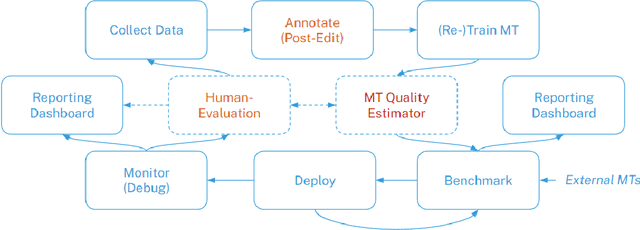
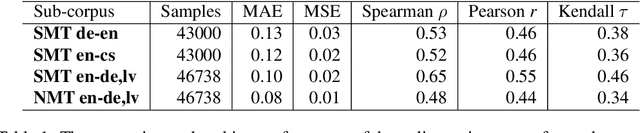
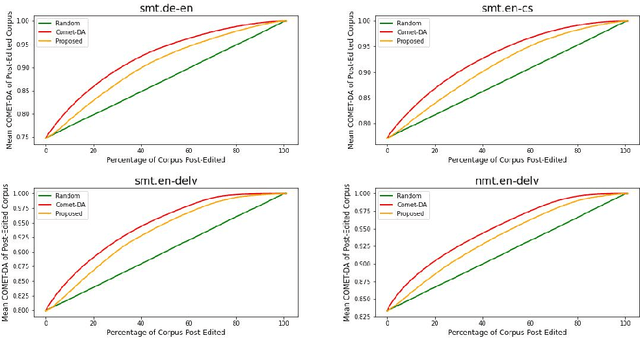
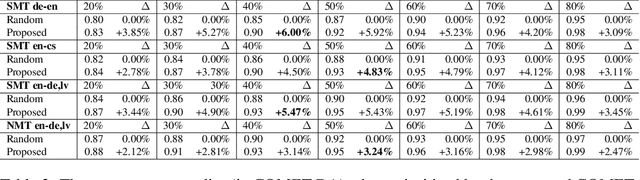
Abstract:This paper proposes an efficient and semi-automated method for human-in-the-loop post-editing for machine translation (MT) corpus generation. The method is based on online training of a custom MT quality estimation metric on-the-fly as linguists perform post-edits. The online estimator is used to prioritize worse hypotheses for post-editing, and auto-close best hypotheses without post-editing. This way, significant improvements can be achieved in the resulting quality of post-edits at a lower cost due to reduced human involvement. The trained estimator can also provide an online sanity check mechanism for post-edits and remove the need for additional linguists to review them or work on the same hypotheses. In this paper, the effect of prioritizing with the proposed method on the resulting MT corpus quality is presented versus scheduling hypotheses randomly. As demonstrated by experiments, the proposed method improves the lifecycle of MT models by focusing the linguist effort on production samples and hypotheses, which matter most for expanding MT corpora to be used for re-training them.
BioFLAIR: Pretrained Pooled Contextualized Embeddings for Biomedical Sequence Labeling Tasks
Aug 13, 2019
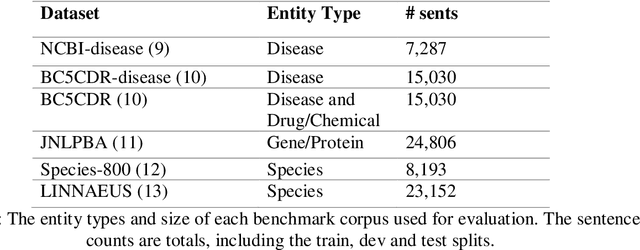
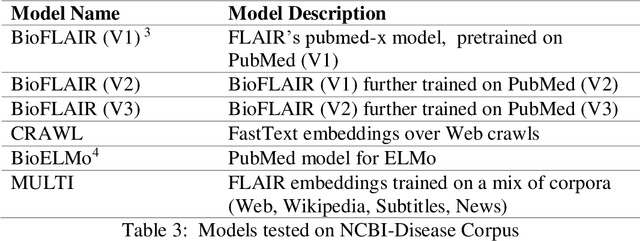
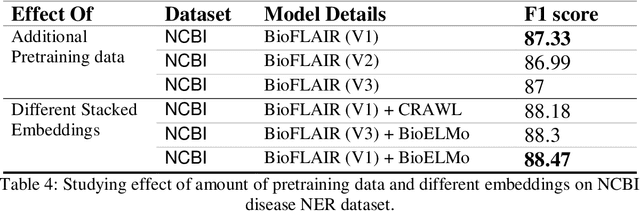
Abstract:Biomedical Named Entity Recognition (NER) is a challenging problem in biomedical information processing due to the widespread ambiguity of out of context terms and extensive lexical variations. Performance on bioNER benchmarks continues to improve due to advances like BERT, GPT, and XLNet. FLAIR (1) is an alternative embedding model which is less computationally intensive than the others mentioned. We test FLAIR and its pretrained PubMed embeddings (which we term BioFLAIR) on a variety of bio NER tasks and compare those with results from BERT-type networks. We also investigate the effects of a small amount of additional pretraining on PubMed content, and of combining FLAIR and ELMO models. We find that with the provided embeddings, FLAIR performs on-par with the BERT networks - even establishing a new state of the art on one benchmark. Additional pretraining did not provide a clear benefit, although this might change with even more pretraining being done. Stacking the FLAIR embeddings with others typically does provide a boost in the benchmark results.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge